SNT1 is ImageJ’s framework for tracing, visualization, quantitative analyses, and modeling of neuronal morphology. For tracing, SNT supports modern multidimensional microscopy data, semi-automated and automated routines, and options for editing traces. For data analysis, SNT features advanced visualization tools, access to all major morphology databases, and support for whole-brain circuitry data.
SNT is based on publications. Please cite SNT in your own research!
Overview
SNT is a toolbox for tracing and analyzing neuronal morphologies imaged using light microscopy. It aims to be as complete as possible, while remaining efficient to use. It can be used as a desktop application, or a scripting library. It supersedes the original Simple Neurite Tracer software, and aggregates other tools previously scattered across the Fiji ecosystem of plugins, including Sholl2 and Strahler plugins.
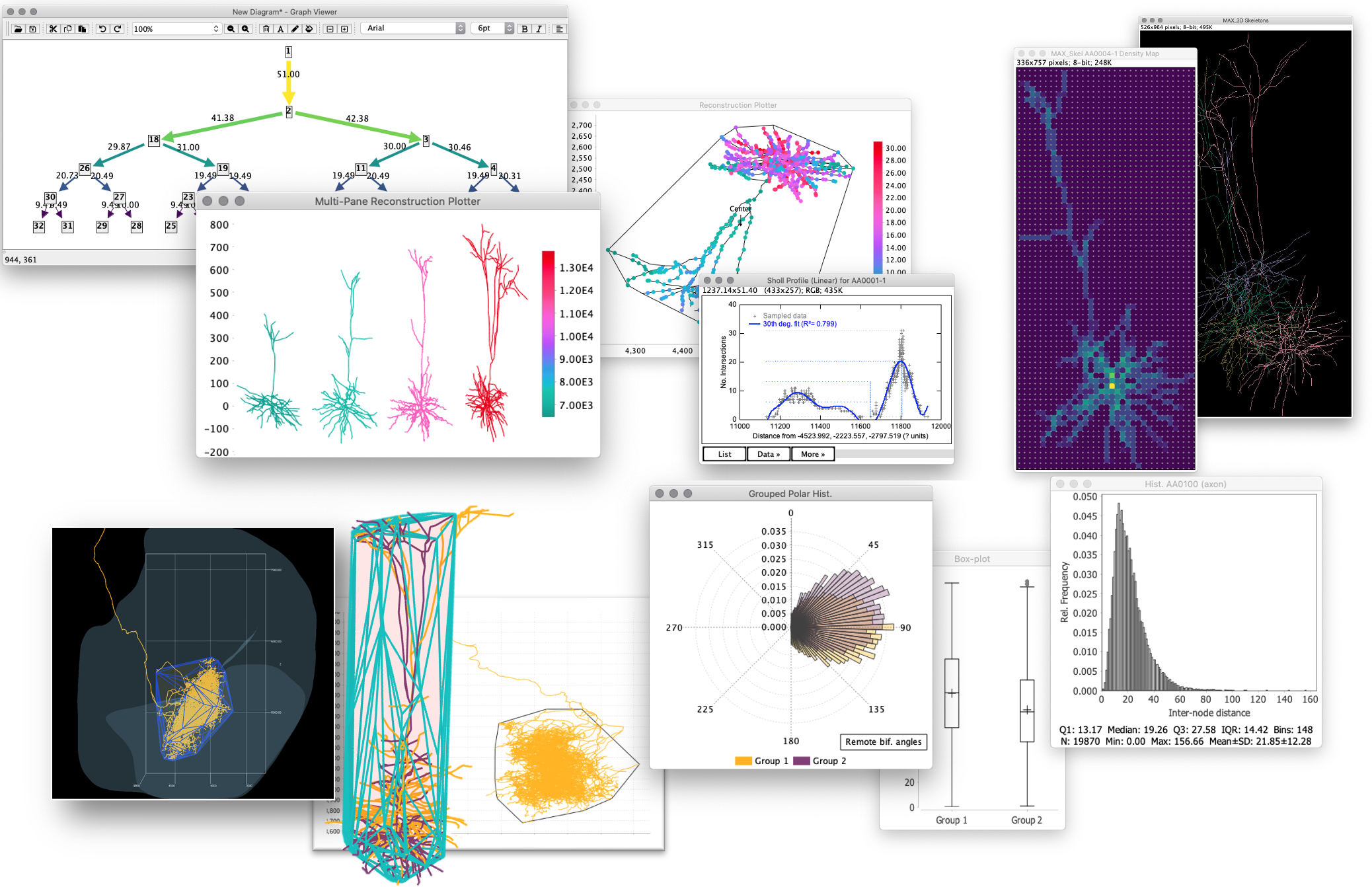
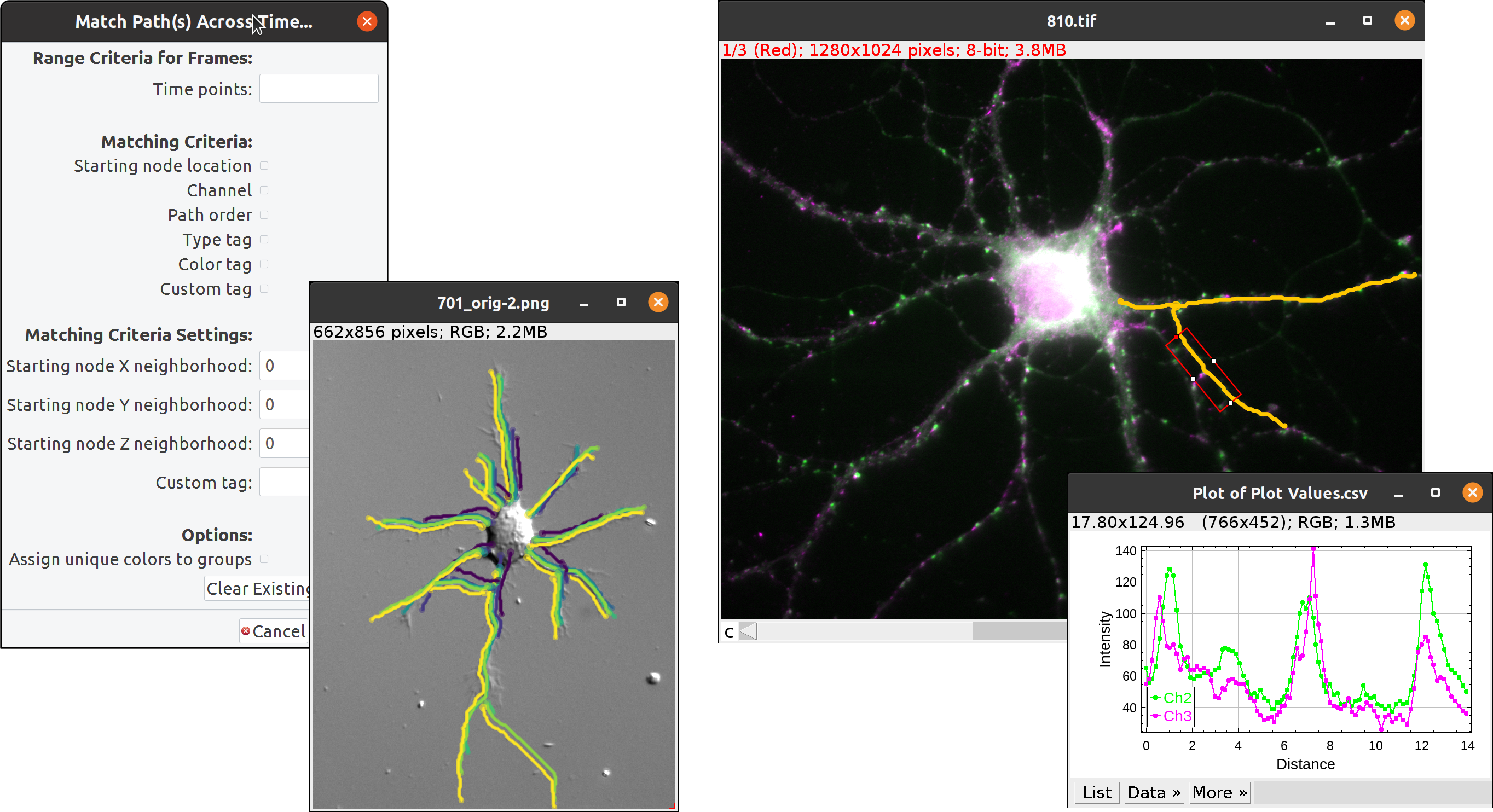
The source repository contains more details about the project, including a list of features, and implementation details. Below is a gallery showcasing SNT’s functionality:
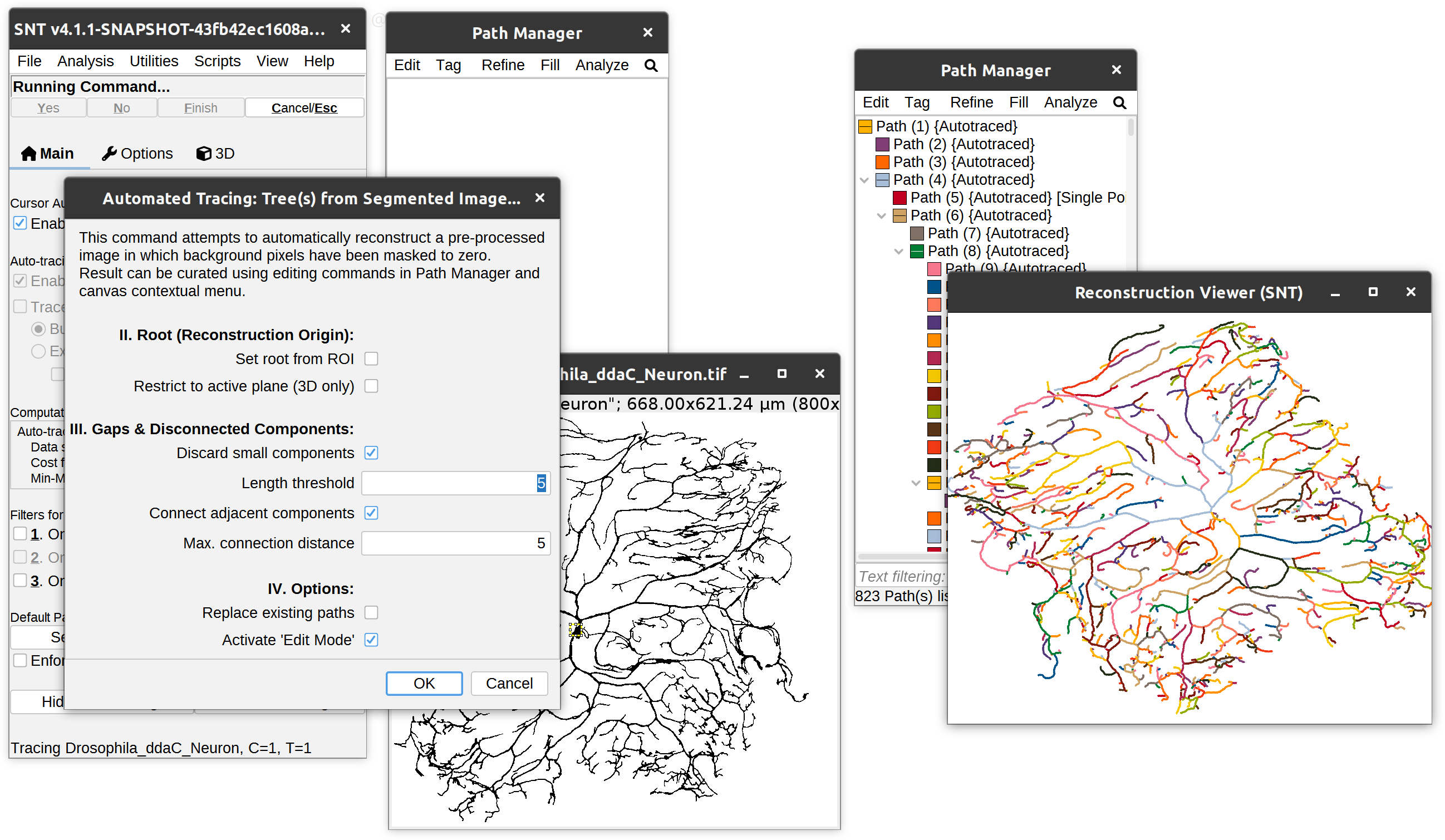 Fully automated tracing from both grayscale images (no thresholding required) and segmented images
Fully automated tracing from both grayscale images (no thresholding required) and segmented images
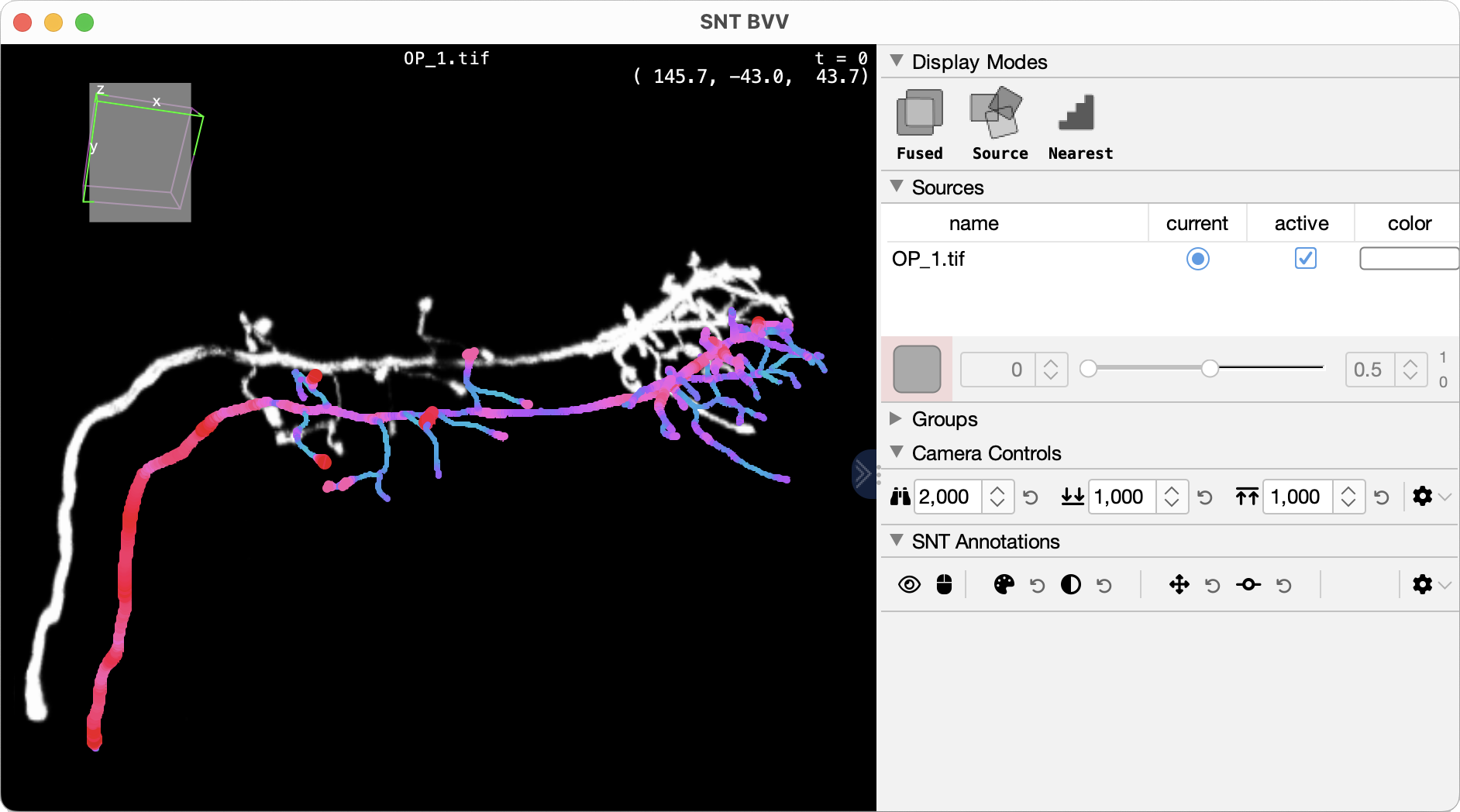 Big-data (out-of-core) support: Automatic switch to disk-backed processing for TB-scale datasets that don’t fit in RAM (experimental)
Big-data (out-of-core) support: Automatic switch to disk-backed processing for TB-scale datasets that don’t fit in RAM (experimental)
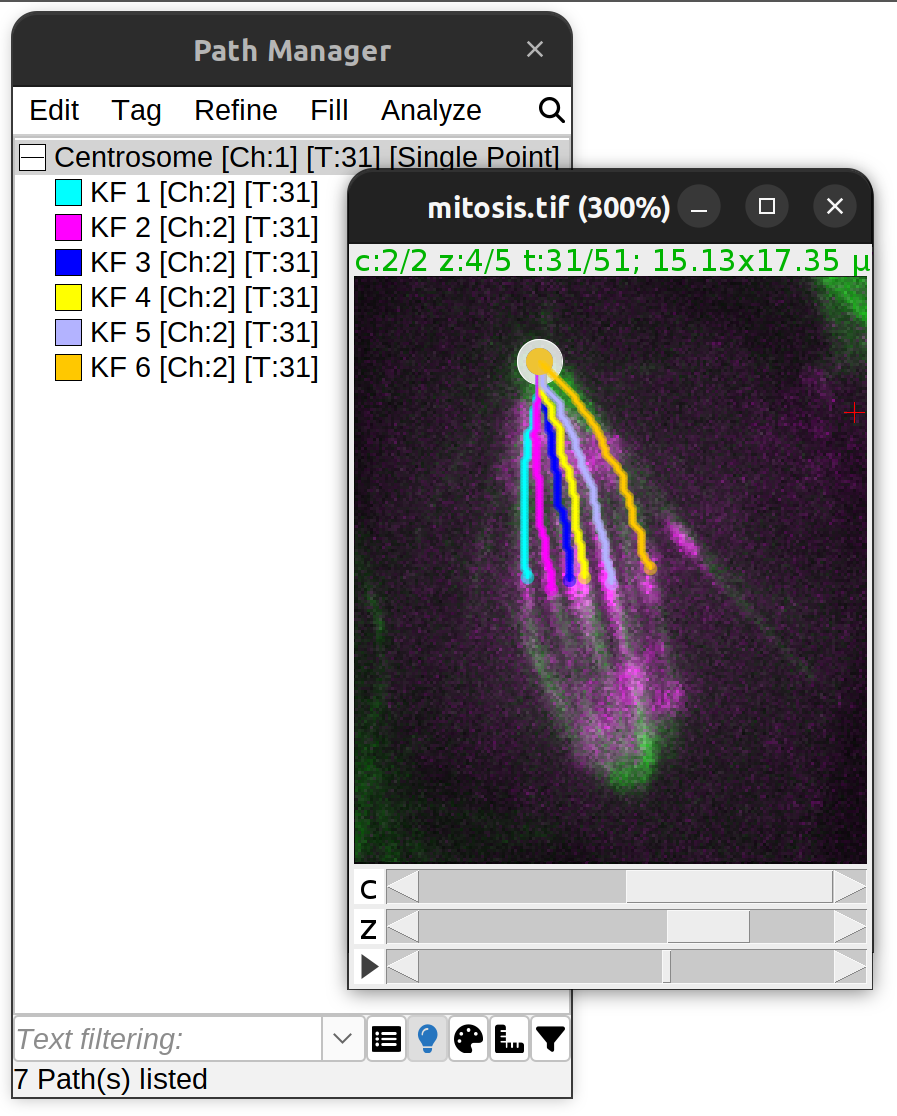
 Semi-automated tracing: Support for multi-channel and timelapse images
Semi-automated tracing: Support for multi-channel and timelapse images
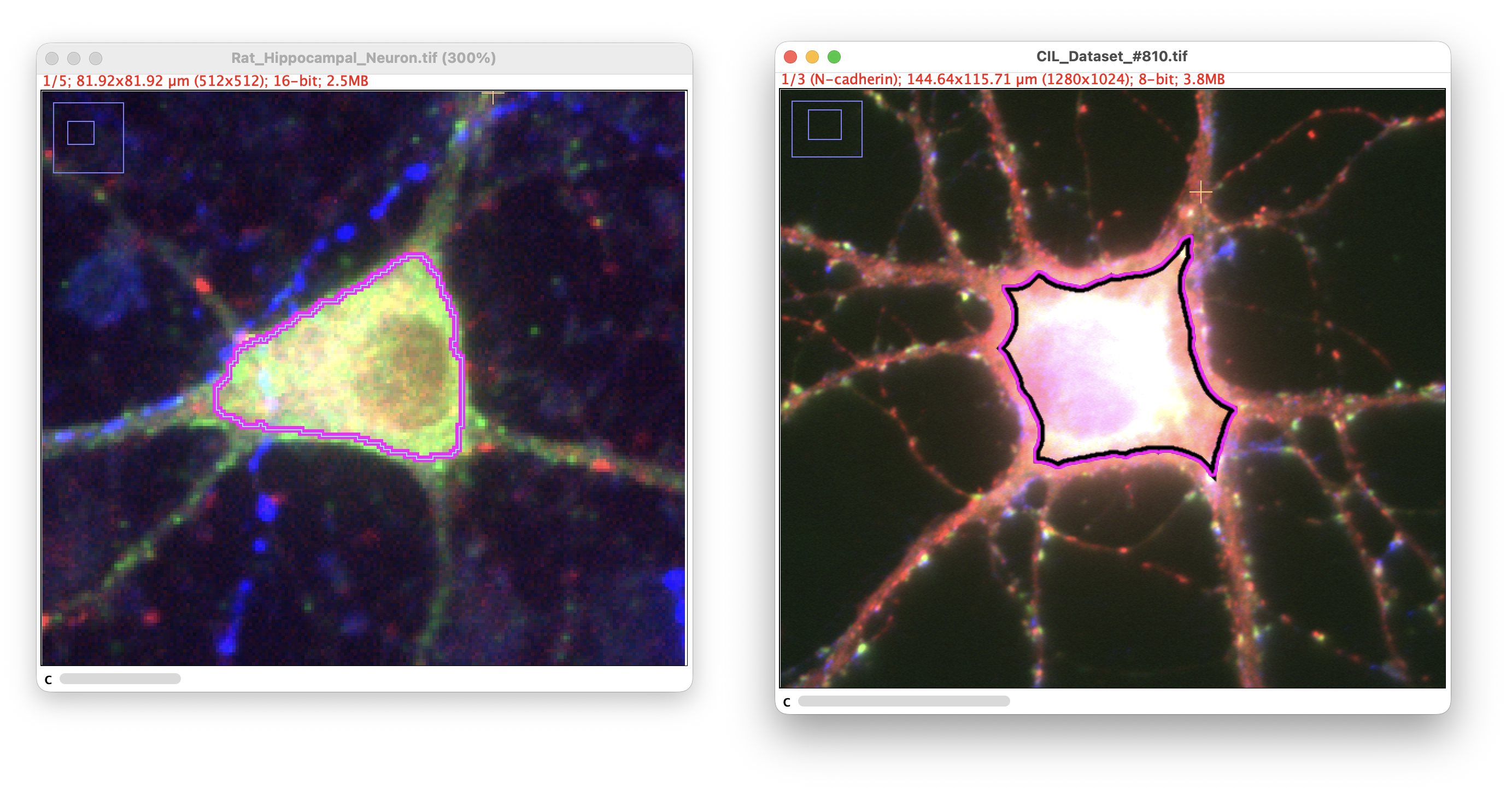 Automated soma detection (no thresholding required)
Automated soma detection (no thresholding required)
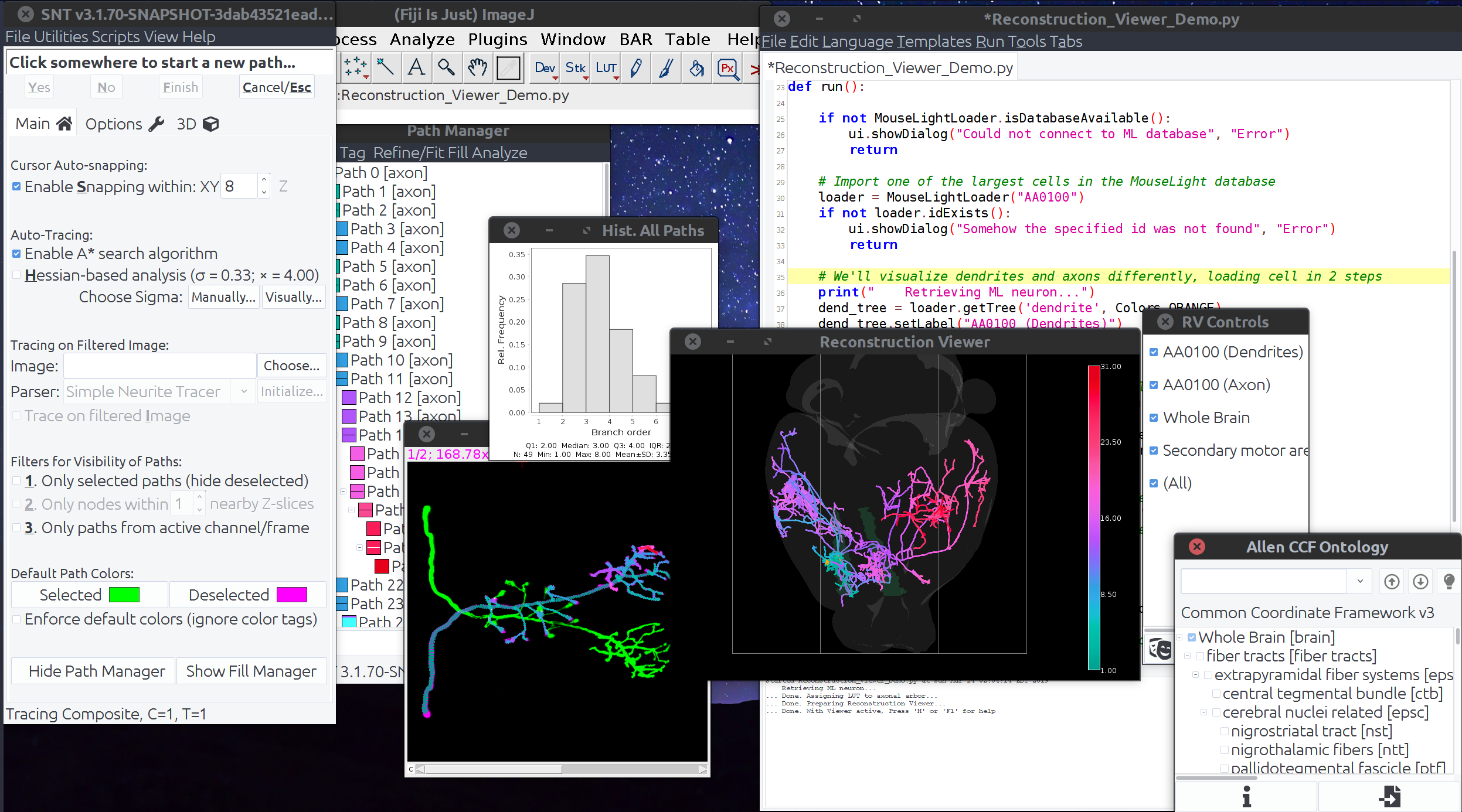 Scripted routines co-exist with graphical user interface operations
Scripted routines co-exist with graphical user interface operations
 Scripting in any of Fiji’s supported languages facilitated by SNT’s Script Recorder
Scripting in any of Fiji’s supported languages facilitated by SNT’s Script Recorder
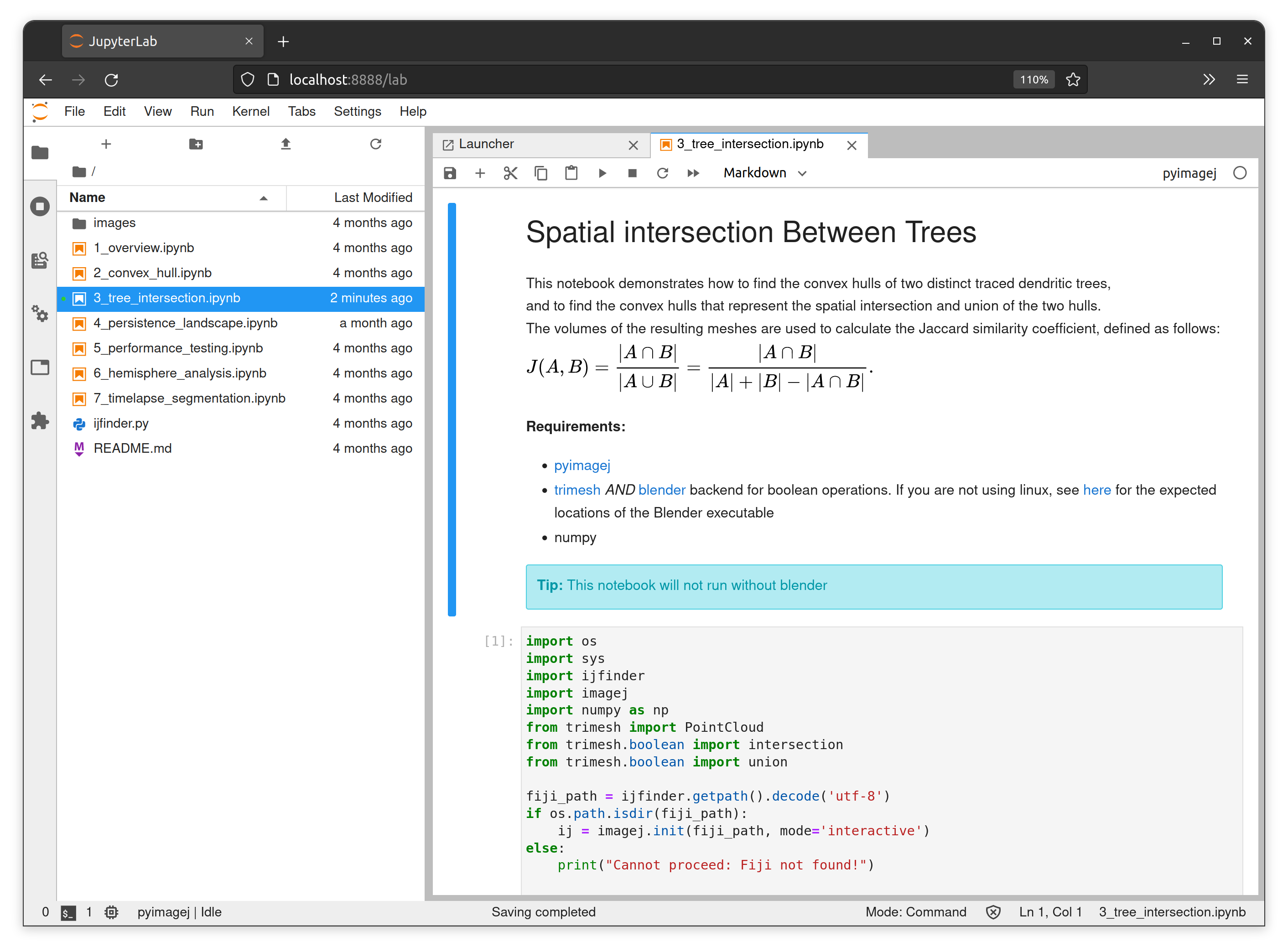 Scripting in native Python using PySNT
Scripting in native Python using PySNT
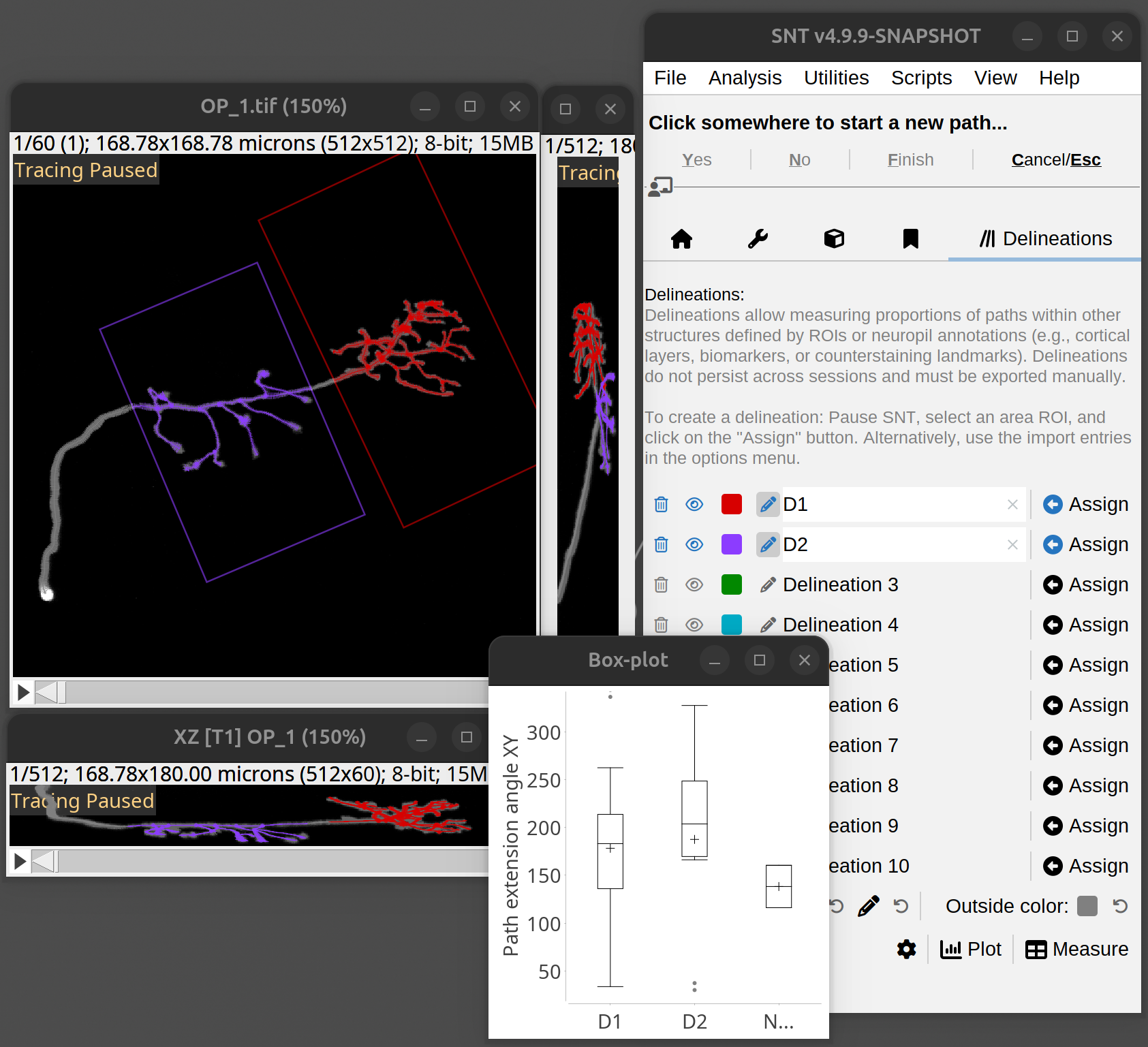
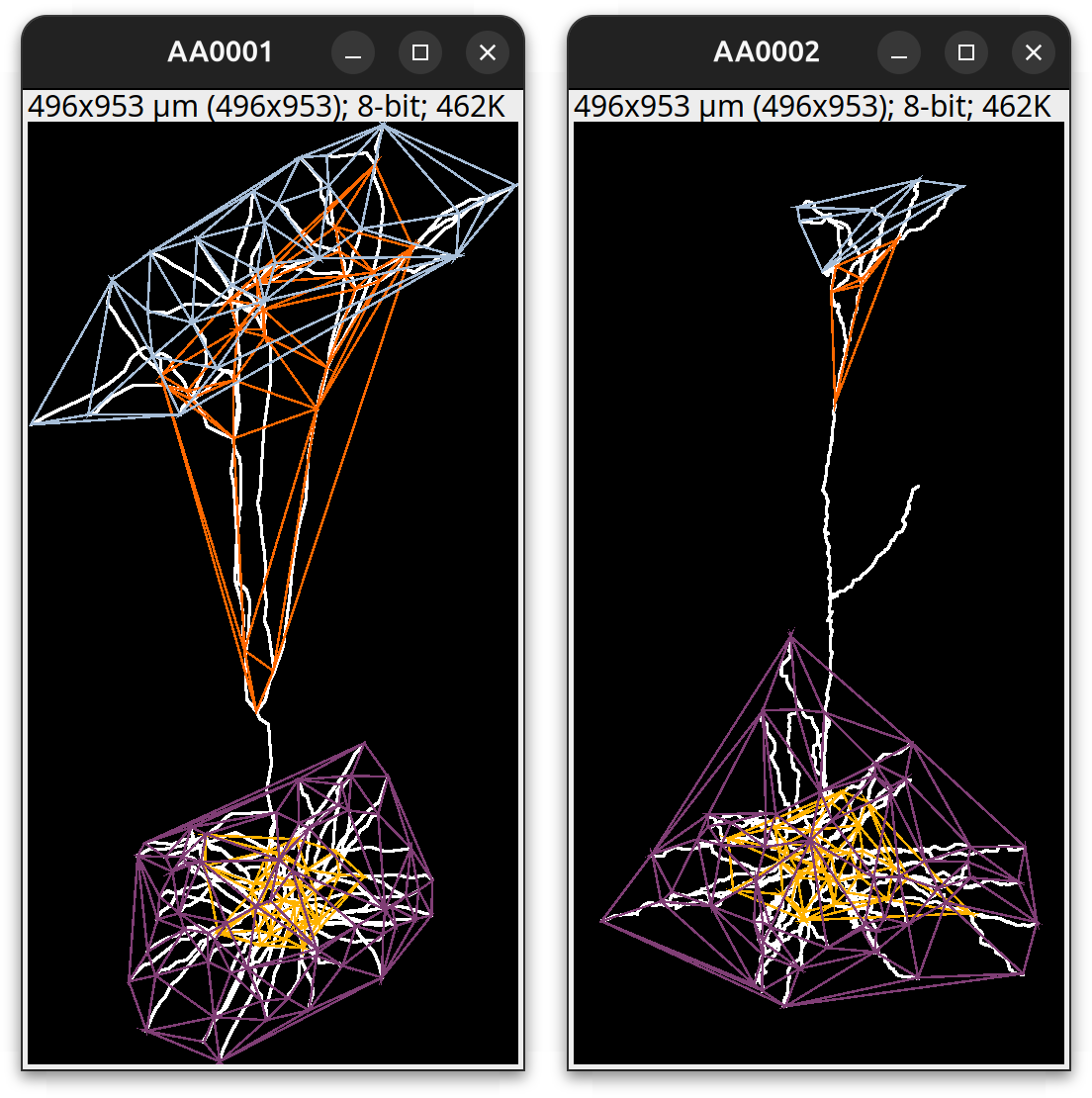 Delaunay tessellation: Tracings can be used in image processing routines
Delaunay tessellation: Tracings can be used in image processing routines
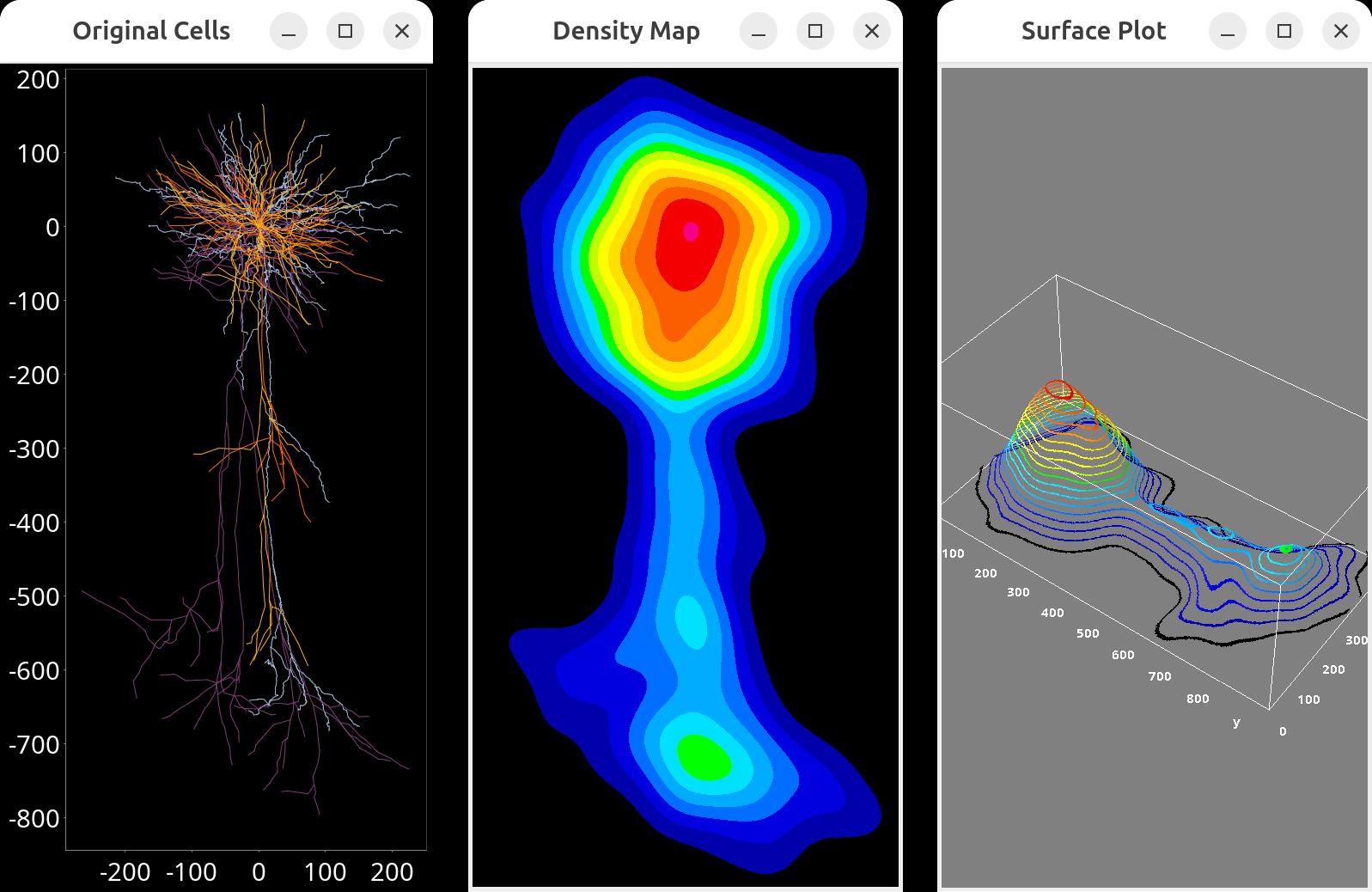 Density maps for group(s) of cells obtained from built-in scripts
Density maps for group(s) of cells obtained from built-in scripts
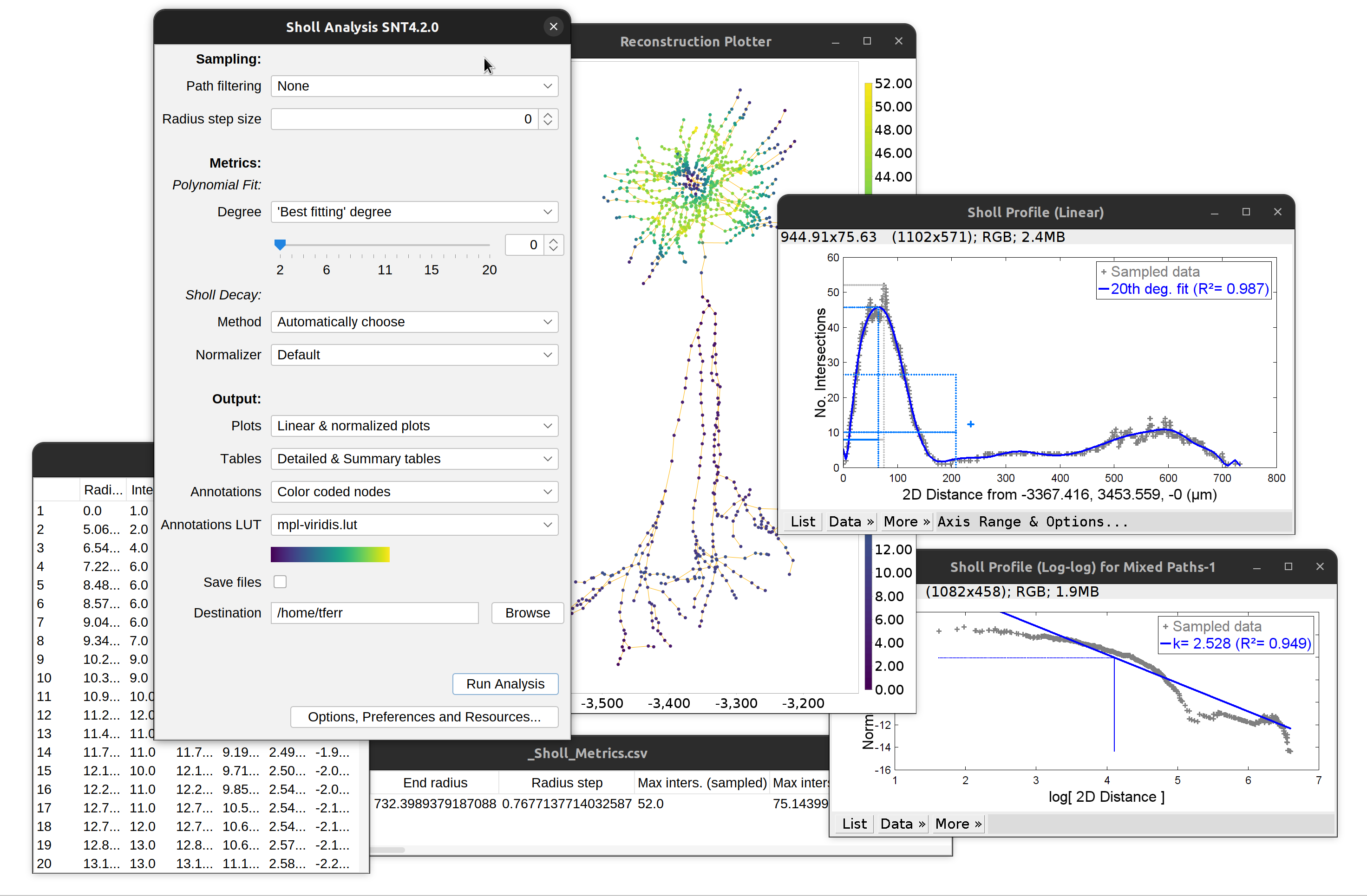
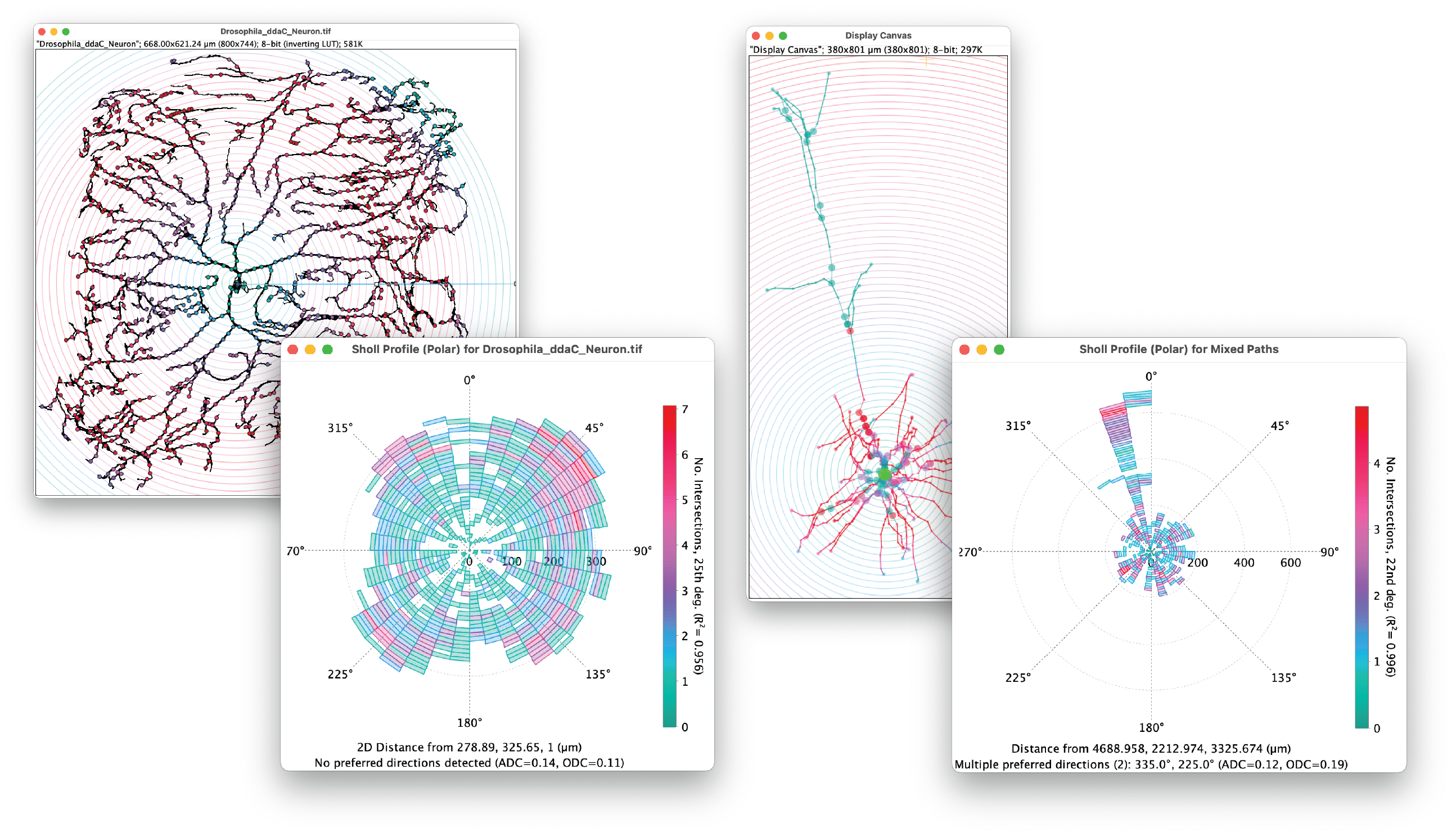 Advanced Sholl-based quantifications
Advanced Sholl-based quantifications
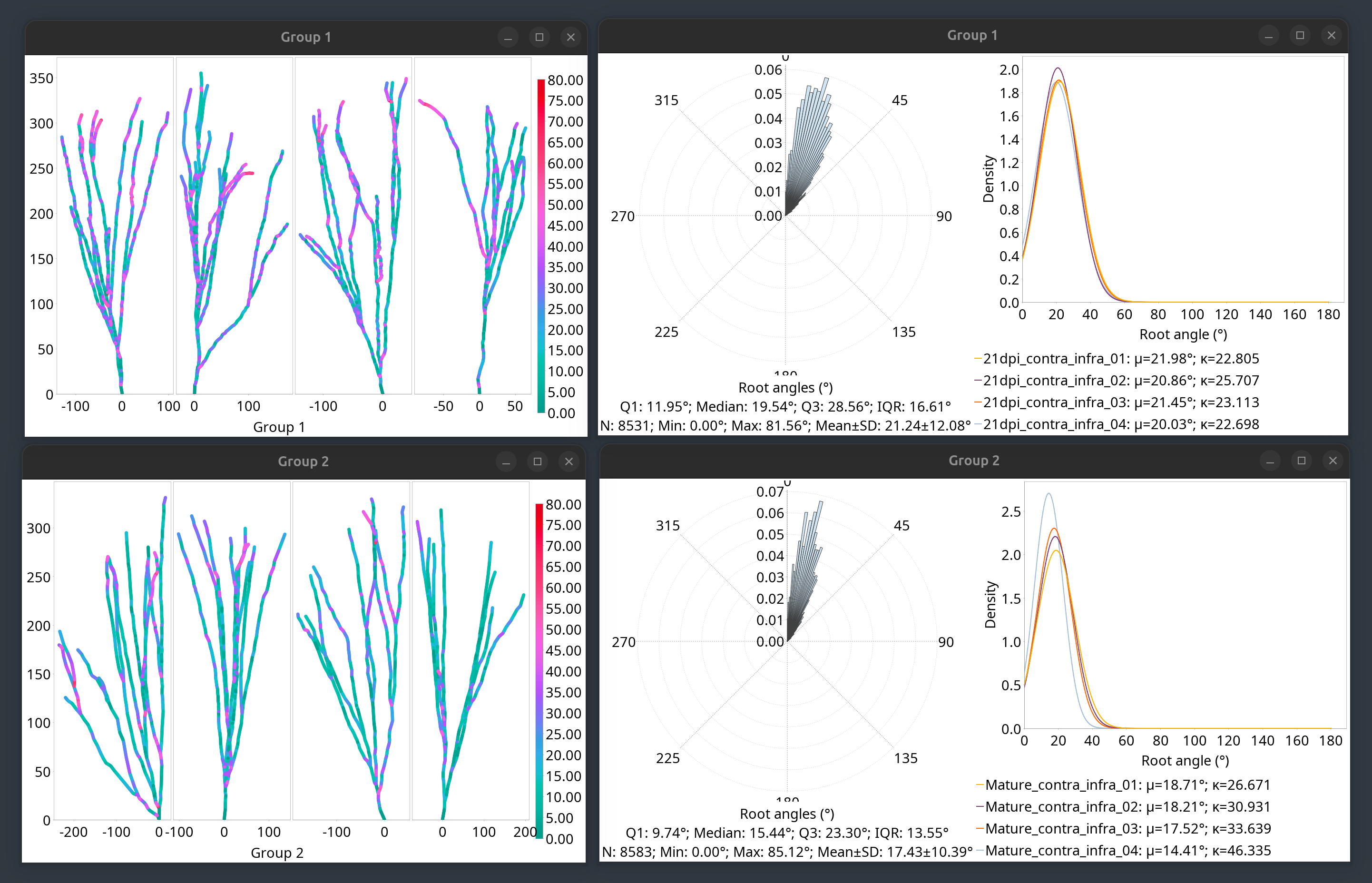
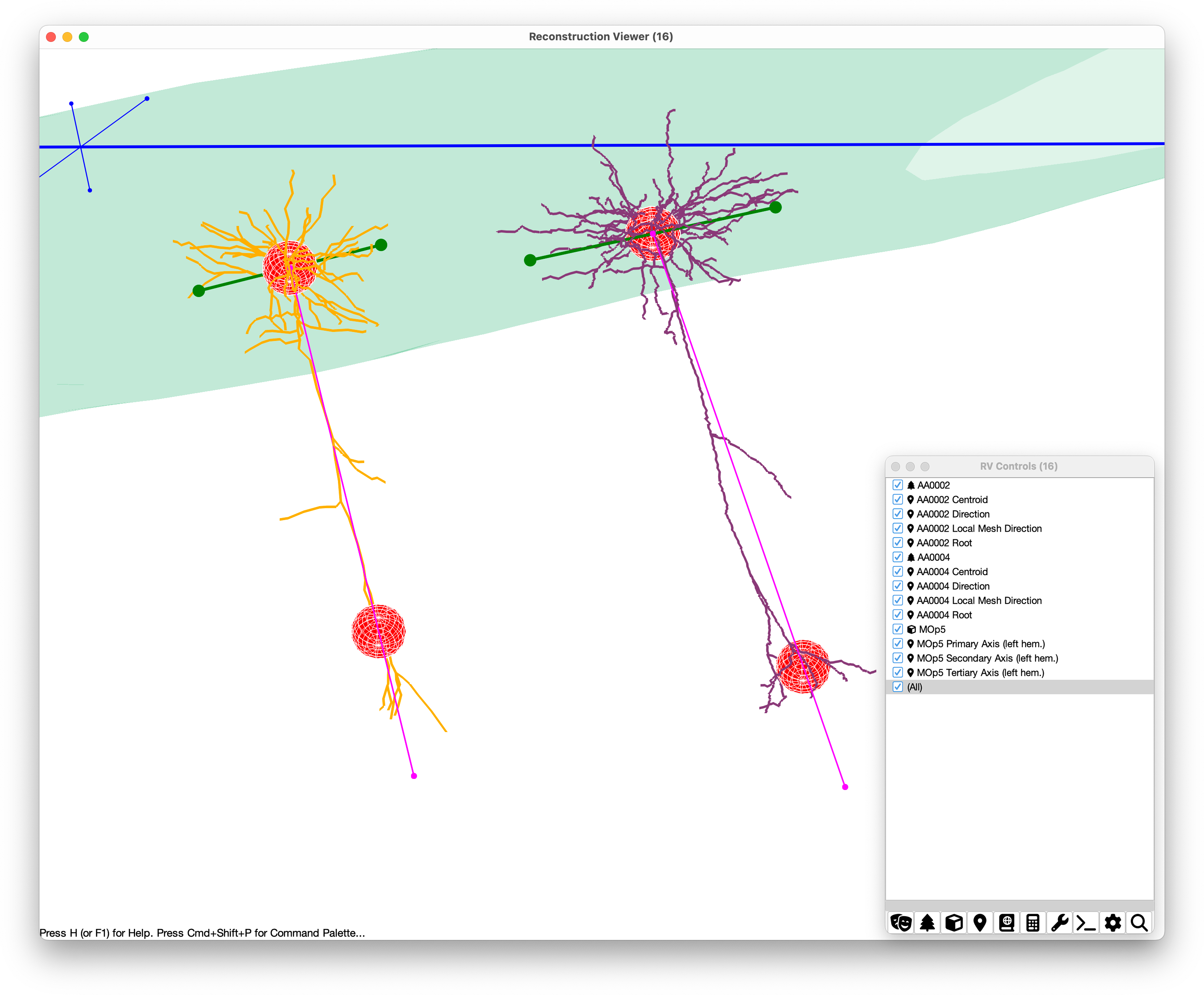 Analysis of surface angles/arbor orientation
Analysis of surface angles/arbor orientation
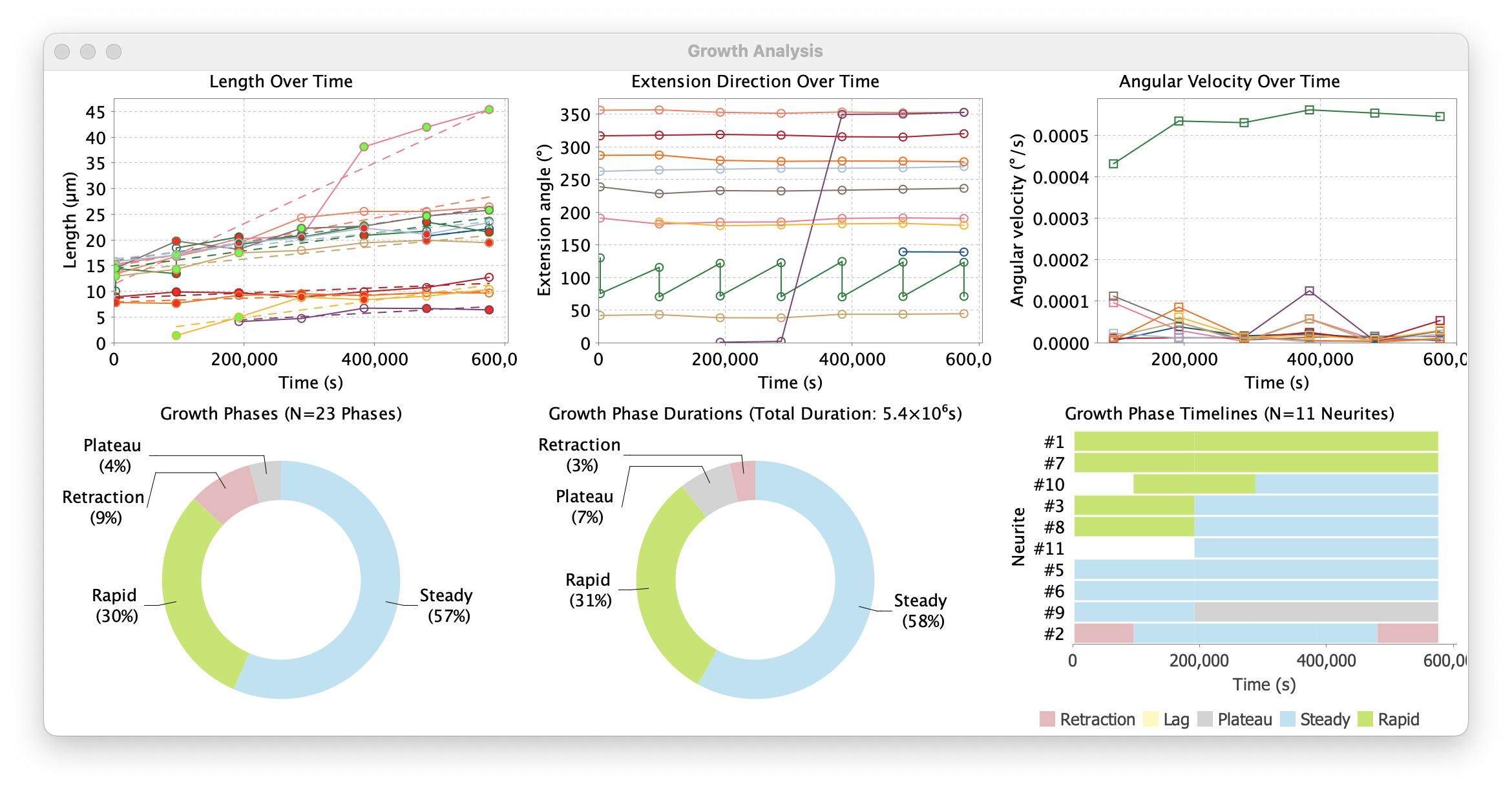
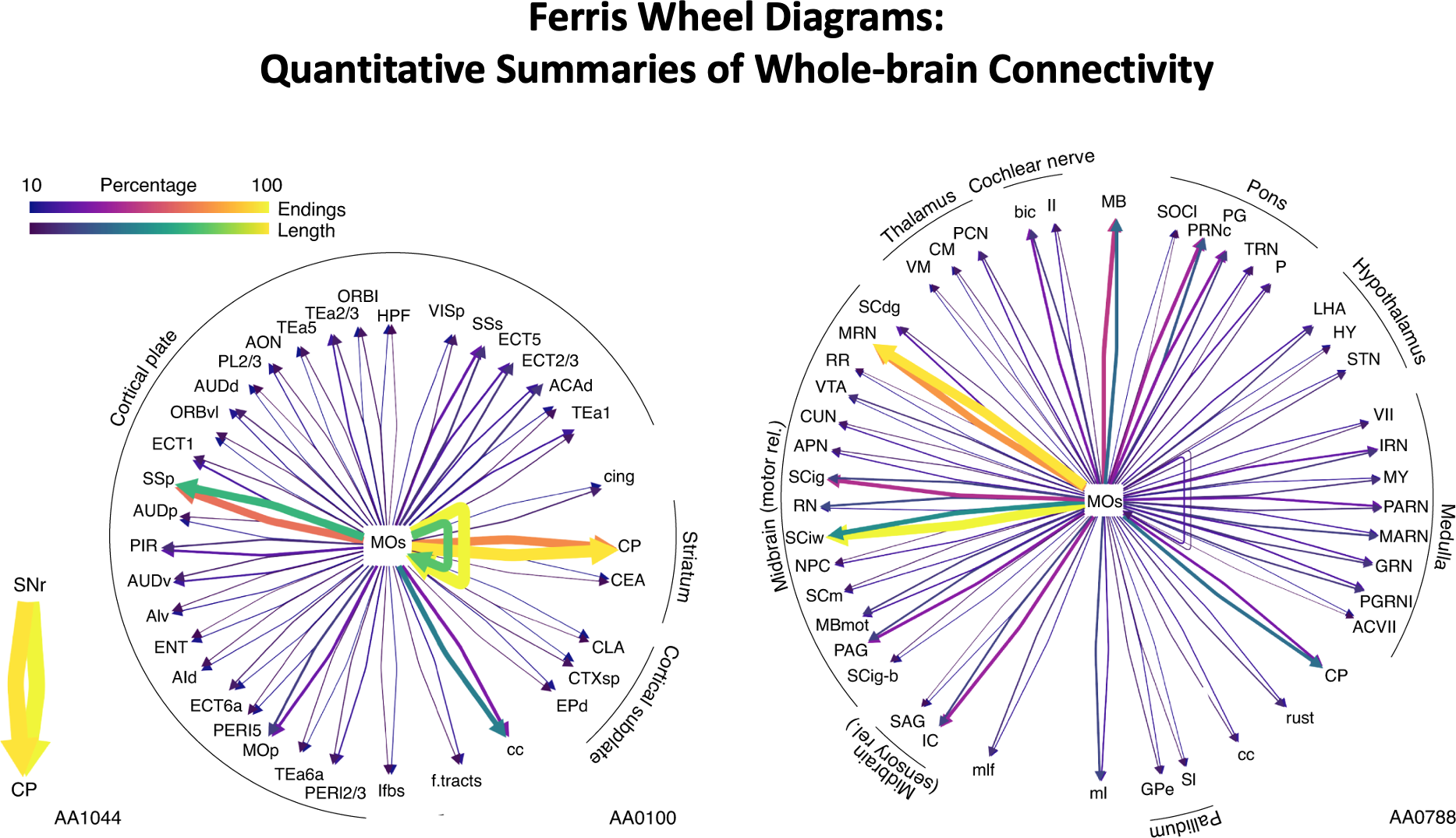 Routines to summarize innervation patterns
Routines to summarize innervation patterns
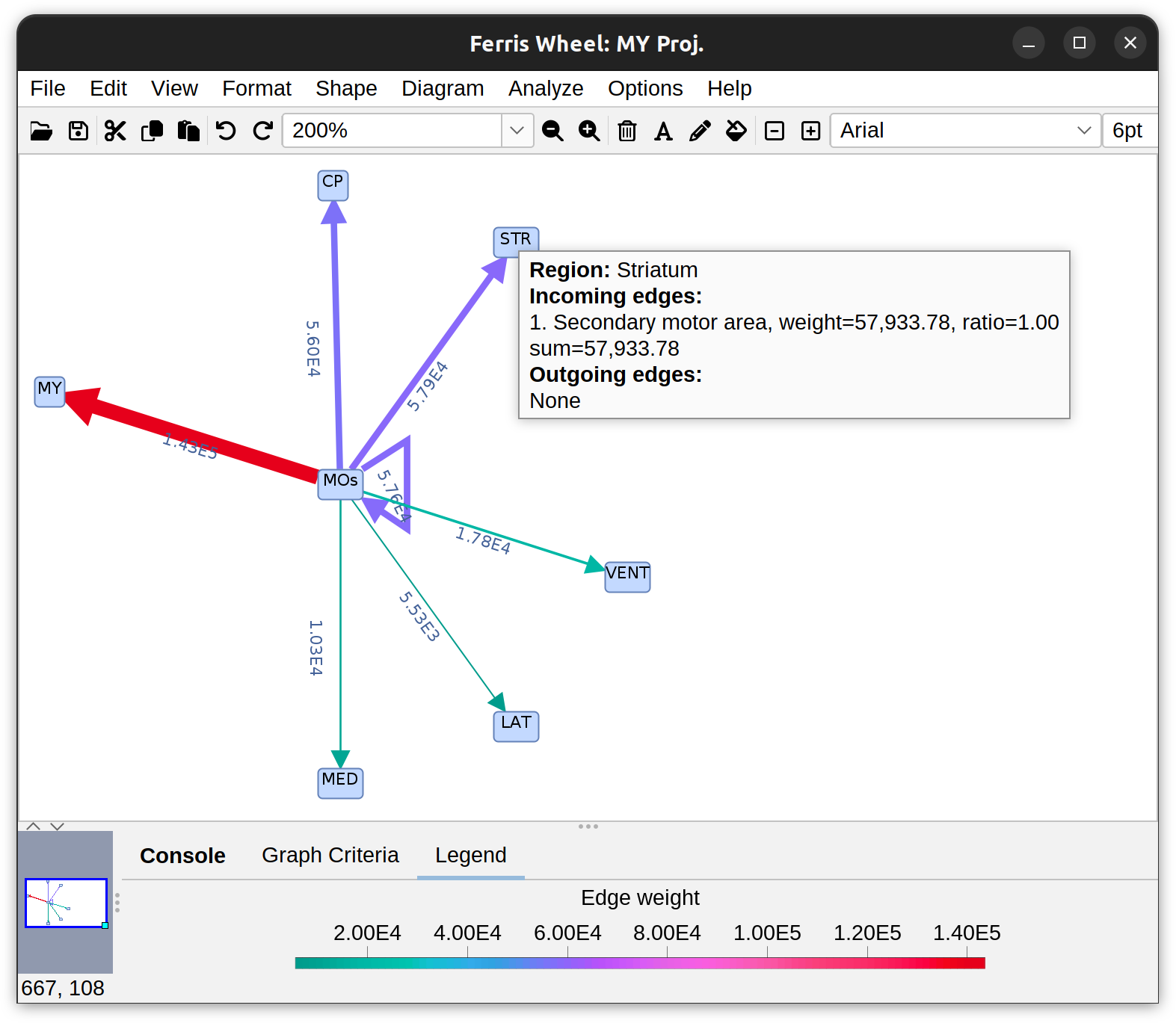 Routines to summarize data from projectomes and connectomics
Routines to summarize data from projectomes and connectomics
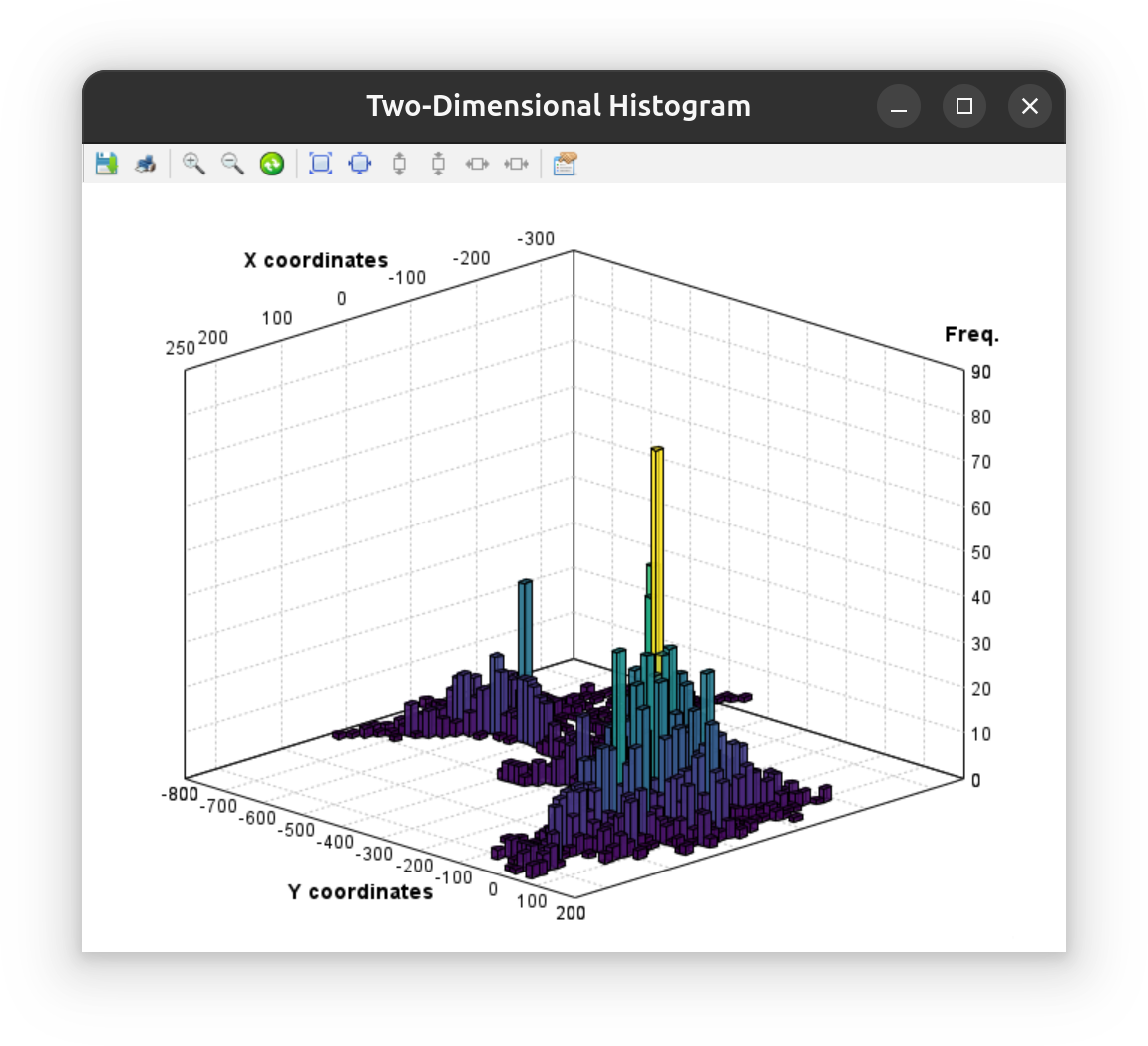 Specialized statistics
Specialized statistics
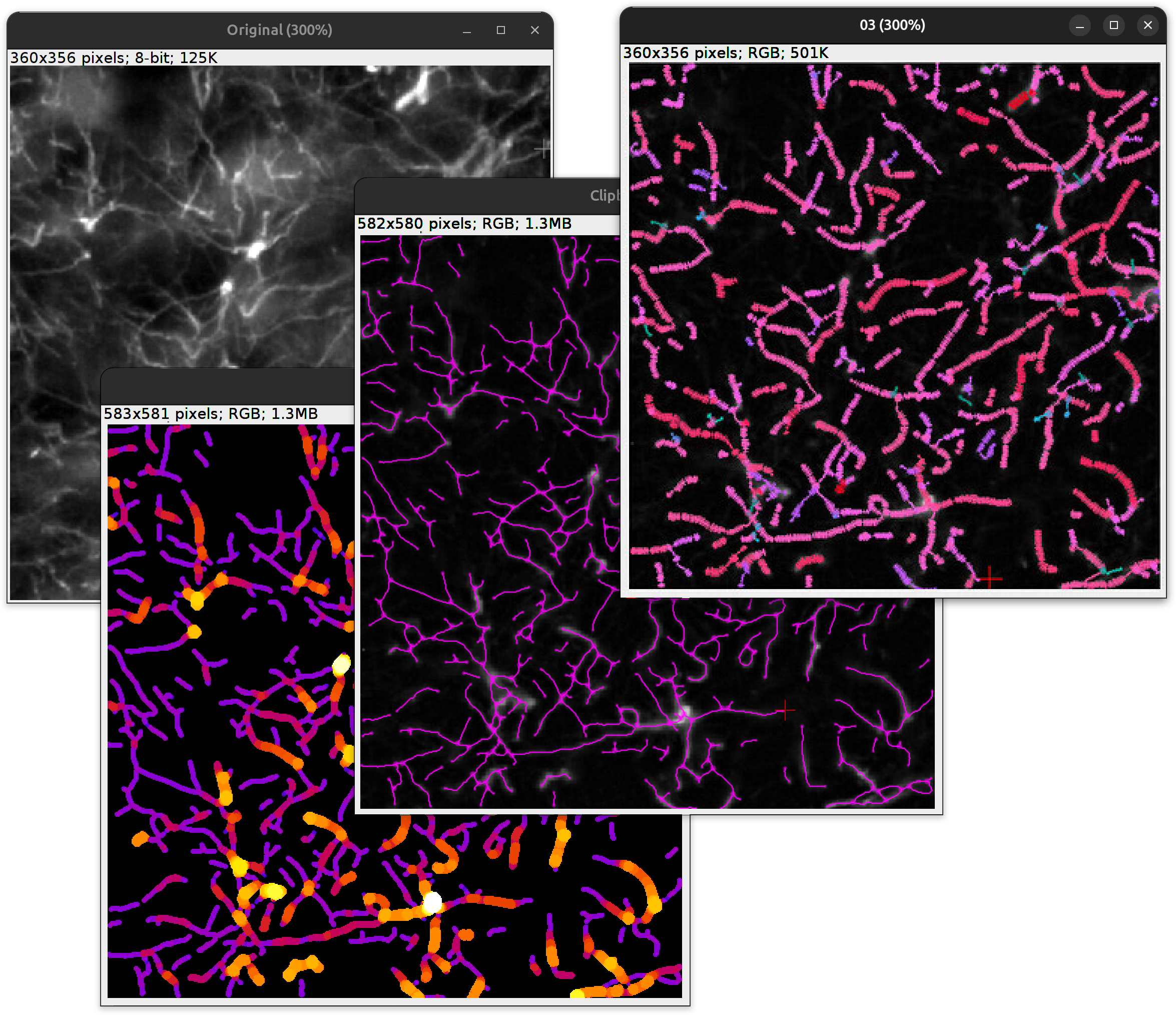 Glia analysis: Bulk characterization (width and length) of astrocytic processes (use case)
Glia analysis: Bulk characterization (width and length) of astrocytic processes (use case)
Installation
SNT requires the latest Fiji version and is distributed through Fiji’s Neuroanatomy update site.
Recommended: Fresh Fiji-Latest Install
- Download Fiji-Latest (bundled with Java 21) from the Fiji downloads page
- Run the Fiji Updater (Help › Update…, the penultimate entry in the Help menu)
- Click Manage update sites
- Search for Neuroanatomy (or SNT) and activate the Neuroanatomy checkbox
- Click Apply changes and restart Fiji
- Optional: For sciview and Cx3D functionality, you need to install sciview. See the official sciview documentation for details.
Upgrading from an older Fiji (Java 8)? SNT no longer supports Java 8. If you are running a legacy Fiji installation, you will see a startup dialog guiding you through the transition. Your existing data and traces are not affected. See the release notes for details on what’s new.
Documentation
SNT’s documentation is extensive. Please use the navigation bar on top of the page to access the different sections, organized as follows:
| Section | Contents |
|---|---|
| Analysis | Overview of GUI-based analyses |
| Autotracing | Documentation on fully automated reconstructions |
| Big Data | Status of big data support |
| Contributing | How to contribute to SNT |
| Extending | Resources for developers interested in extending SNT or parsing TRACES |
| FAQ | Frequently asked questions |
| Key Shortcuts | List of SNT shortcuts (keyboard cheatsheet) |
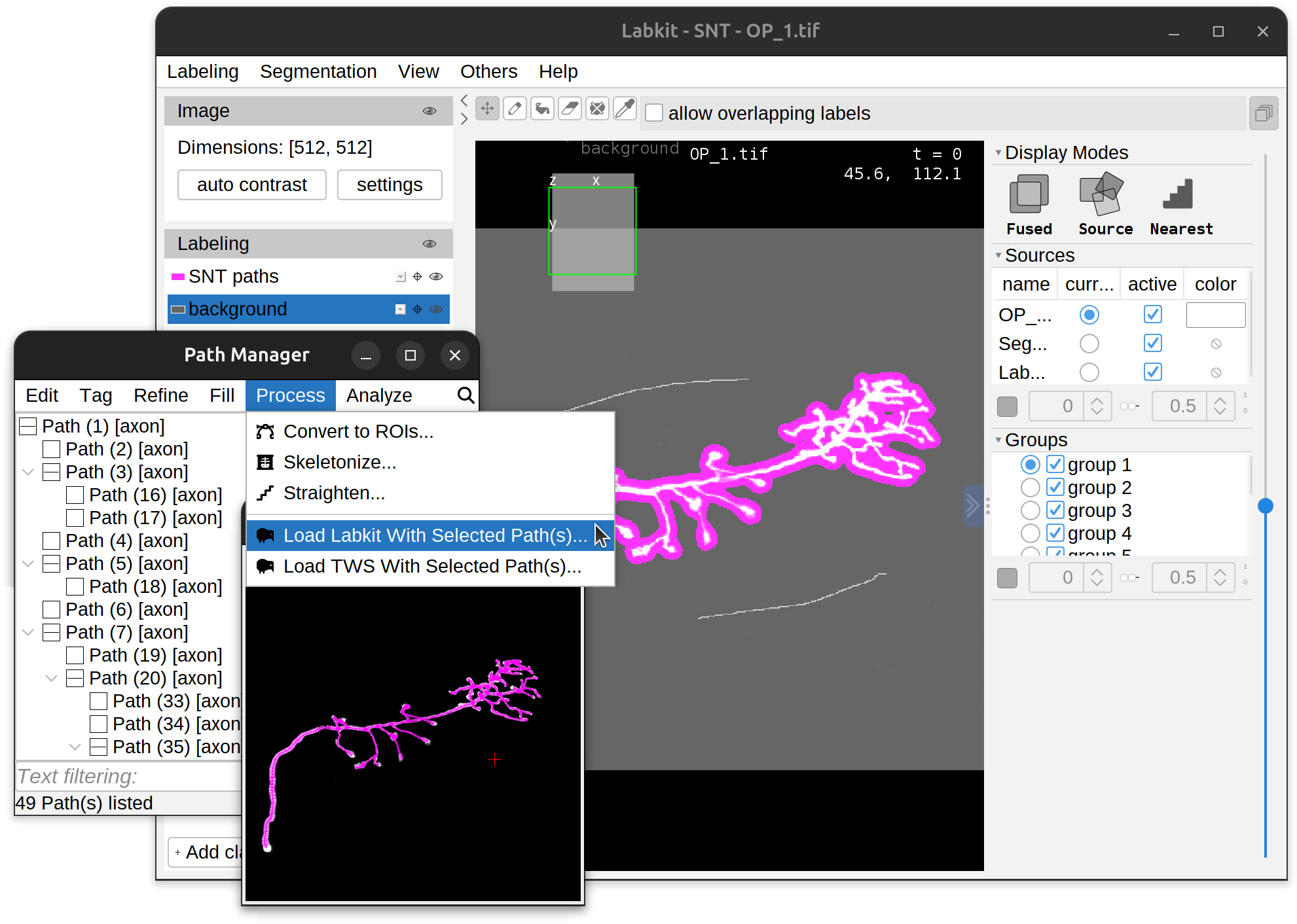
| Machine Learning | Semantic segmentation: Labkit and TWS integration |
| Manual | User guide for main interface and tracing operations |
| Metrics | Definition of metrics |
| Modeling | Cx3D integration |
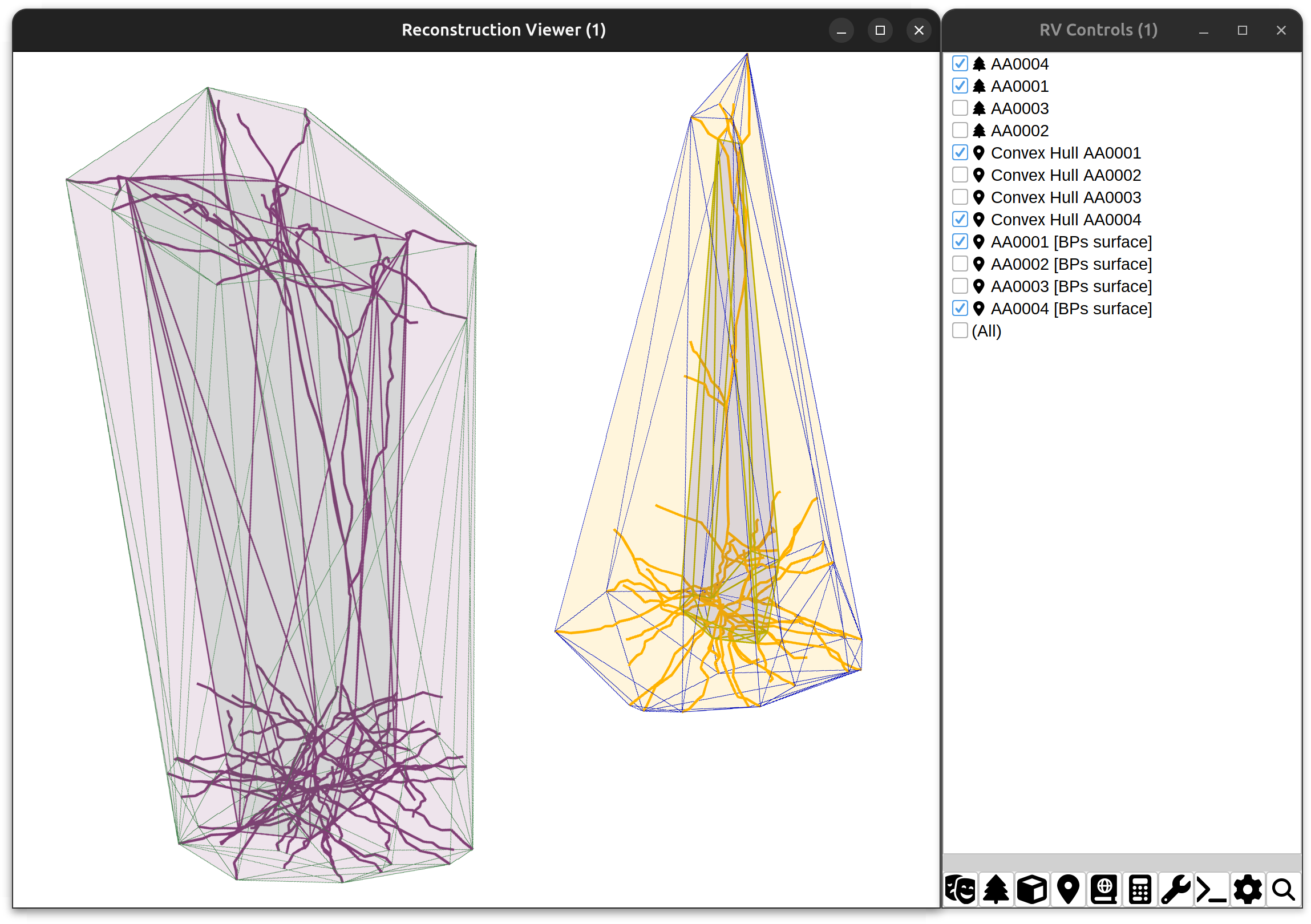
| Reconstruction Viewer | SNT’s entry point for visualization of pre-existing data. If you are analyzing neuronal reconstructions you may want to start here |
| Screencasts | Video tutorials. If you are using SNT for the first time you probably want to start here |
| Scripting | Details on how to use SNT as a scripting library |
| Walkthroughs | Detailed step-by-step instructions for specific tasks |