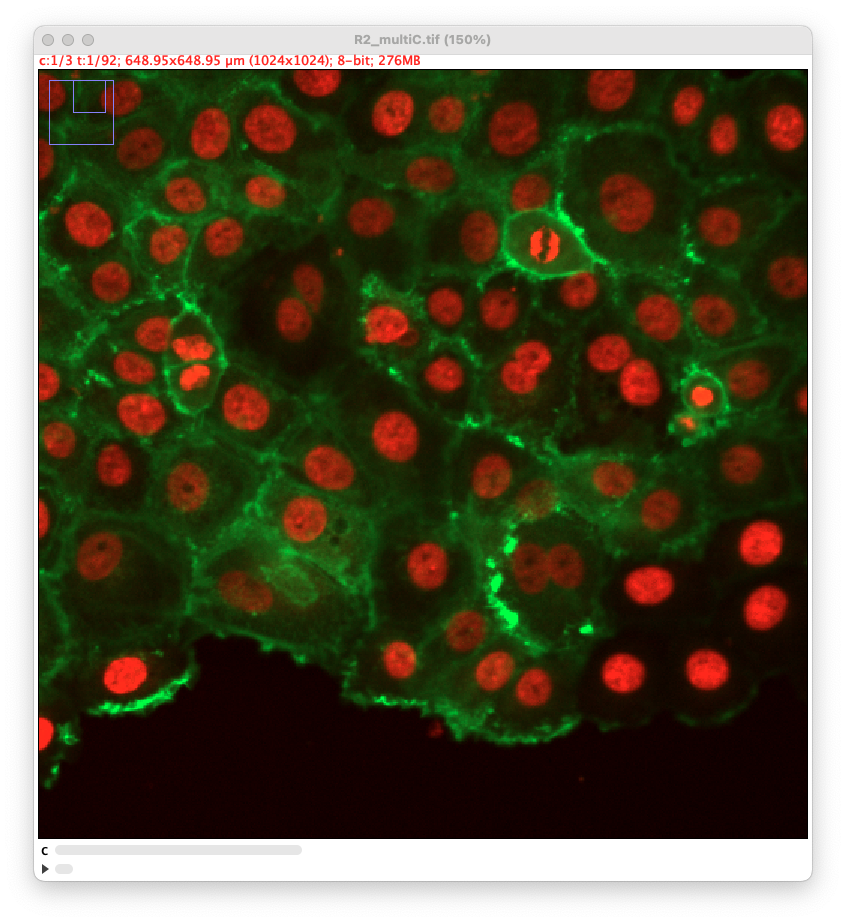
This page describes a detector module for TrackMate that relies on the latest version of cellpose to segment cells in 2D. It is not included in the core of TrackMate and must be installed via its own update site. It also requires cellpose to be installed on your system and working independently. This tutorial page gives installation details and advices at how to use the cellpose integration in TrackMate.
Cellpose-SAM is the latest version of cellpose (at least in August 2025) that leverages a new encoder based on SAM foundational model to achieve better generalizations performance. Compared to the version 3 of cellpose, which integration in TrackMate is documented here and here, Cellpose-SAM has fewer parameters, is more demanding computationally, and generalizes better.
If you use this detector for your research, please also cite the cellpose paper:
Installation
We need to subscribe to an extra update site in Fiji, and have a working installation of cellpose on your system.
TrackMate-Cellpose update site
In Fiji, go to Help › Update…. Update and restart Fiji until it is up-to-date. Then go to the update menu once more, and click on the Manage update sites button, at the bottom-left of the updater window. A new window containing all the known update sites will appear. Click on the TrackMate-Cellpose check box and restart Fiji one more time.
Cellpose-SAM
This step requires you to have a working conda installation, like for any of the Python tools integrated in TrackMate. We recommend miniforge.
For Cellpose specifically, we copy below the installation instruction from the cellpose repository
conda create --name cellpose-sam python=3.10
conda activate cellpose-sam
python -m pip install 'cellpose[gui]'
On Windows, we noticed that it is not enough to get GPU acceleration even if you have a NVIDIA card. You need to reinstall torch with GPU CUDA support, as follow:
- Remove the current version of torch
pip uninstall torch - Reinstall torch with GPU support. Follows the instructions here.
In August 2025, this amounts to:
pip3 install torch torchvision --index-url https://download.pytorch.org/whl/cu126
Then:
cellpose --version
Welcome to CellposeSAM, cellpose v
cellpose version: 4.0.6
platform: win32
python version: 3.12.11
torch version: 2.8.0+cu126
You can ignore the warning if any.
If you have not done it yet, you need to configure the TrackMate conda path in Fiji.
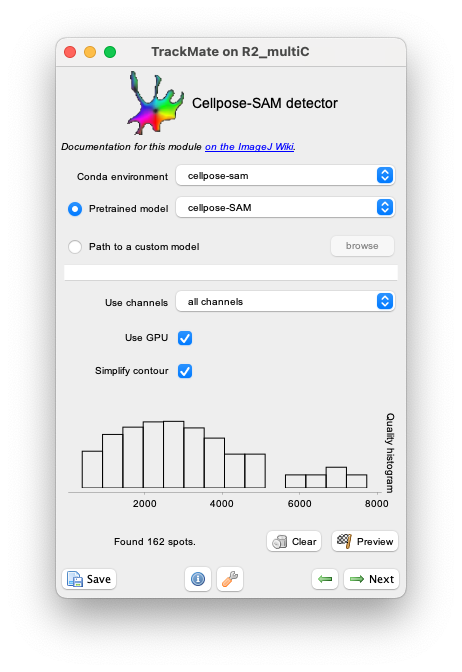
Usage
Cellpose parameters in the TrackMate UI
 We document these parameters from top to bottom in the GUI.
We document these parameters from top to bottom in the GUI.
Conda environment
Specify in what conda environment you installed Cellpose-SAM. If you get an error at this stage, it is likely because the conda path for TrackMate was not configured properly. Check this page.
Pretrained model
Cellpose-SAM has only one pretrained model now: cpsam, which is denoted cellpose-SAM in the UI.
Path to a custom model
You can use a custom cellpose model saved on disk.
First click on the radio button next to the Path to a custom model label, and enter below the path to the model.
It will be used for detection.
Note that the path must be the an absolute path to the file.
Use channels
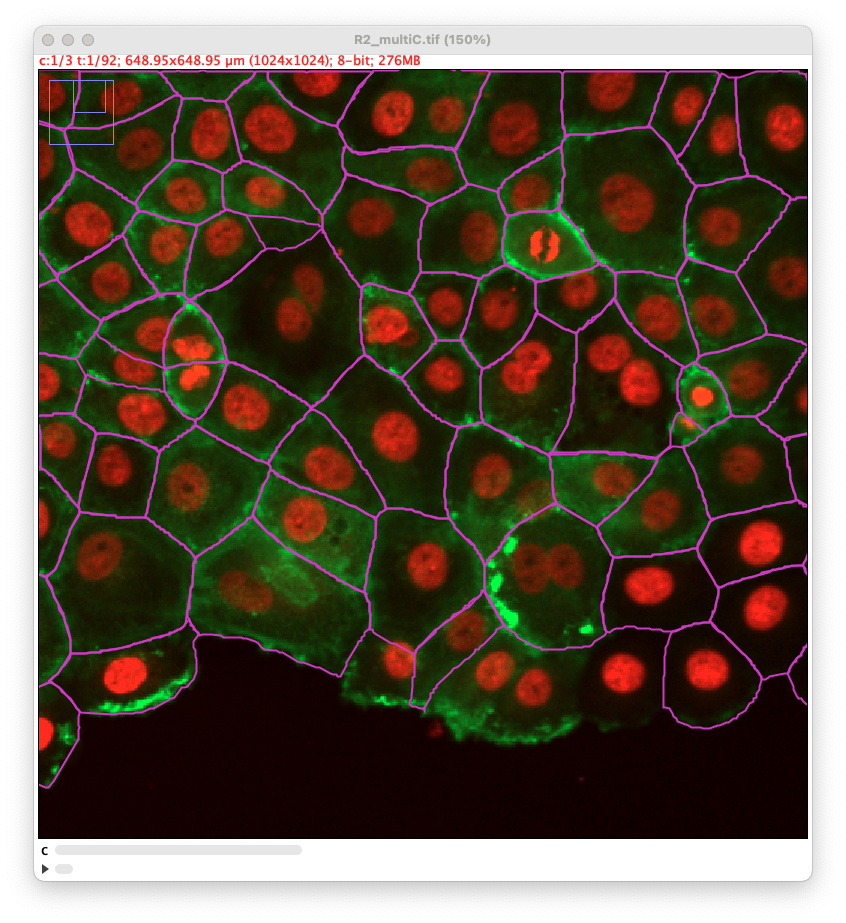
Cellpose-SAM normally disregard the channels parameters that were used up to the version 3. Here however we have added the option to pass to cellpose either the full image with all channels, or just one channel. Depending on the image content, this changes the results quite a lot. This exemplified in the next section.
Use GPU
If this box is checked, the GPU will be used. With modern version of cellpose, even on MacOS, there is not reason not to check this box. Unchecking the box will force cellpose to use the CPU even if GPU support is available, and will process the image with multithreading.
Simplify contours
If this box is checked, the contour outlines generated by the masks returned by cellpose will be smoothed and simplified. This is the same treatment that is used by other TrackMate detectors and documented here.
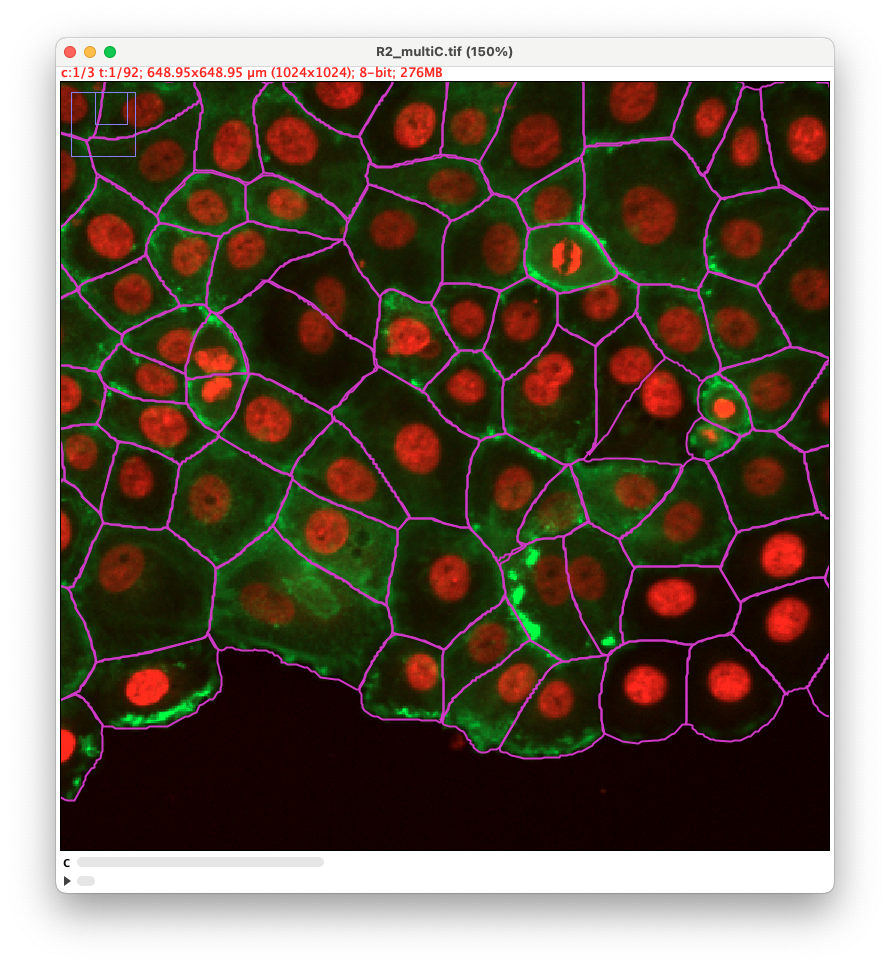
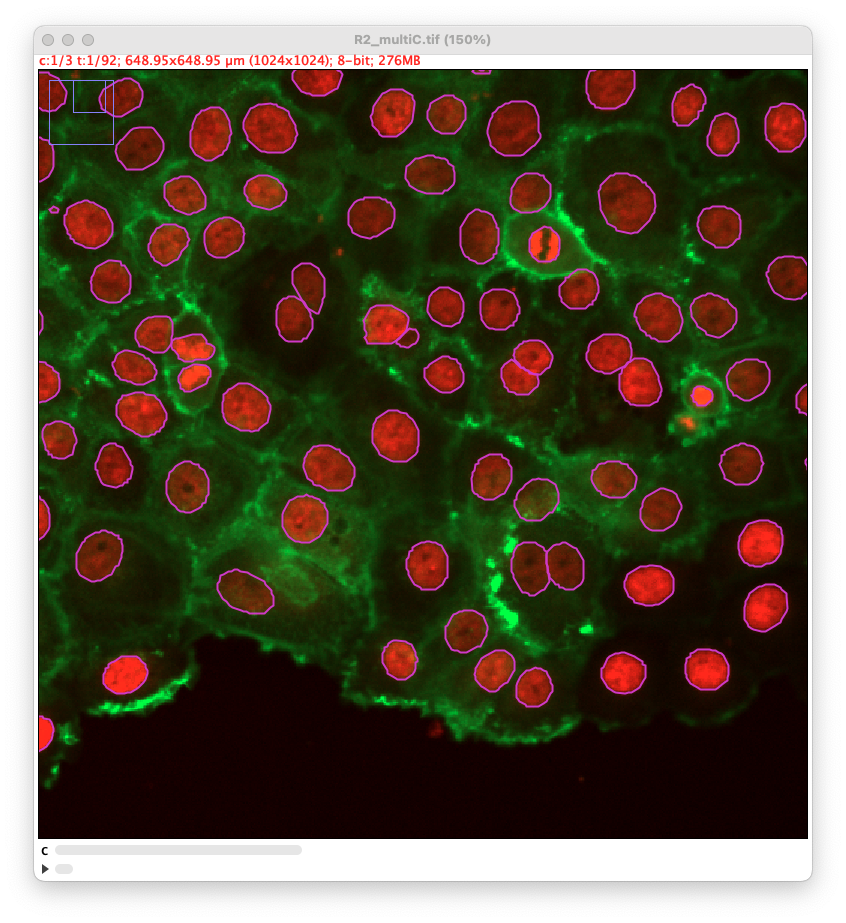
Using different channels
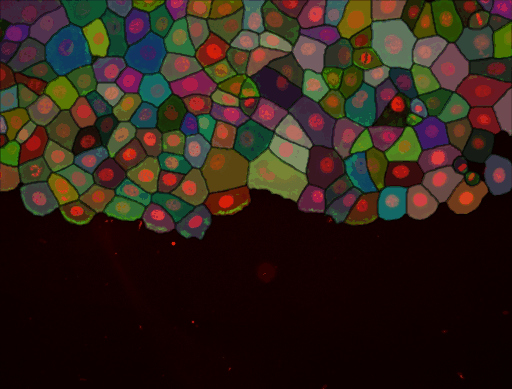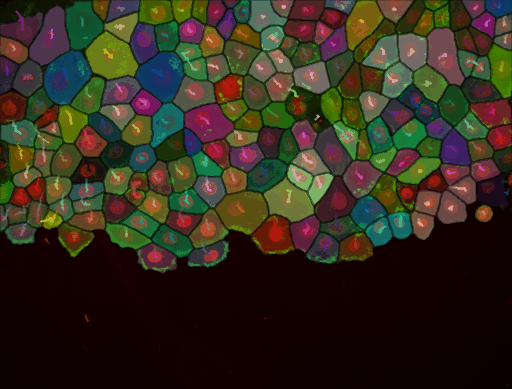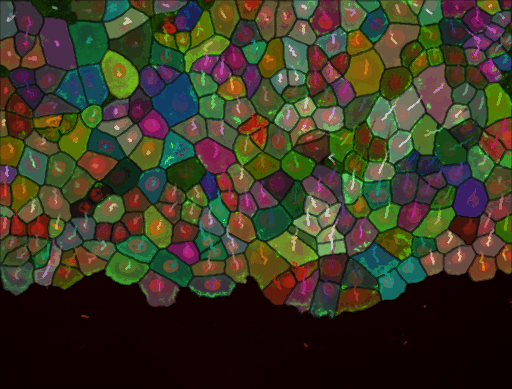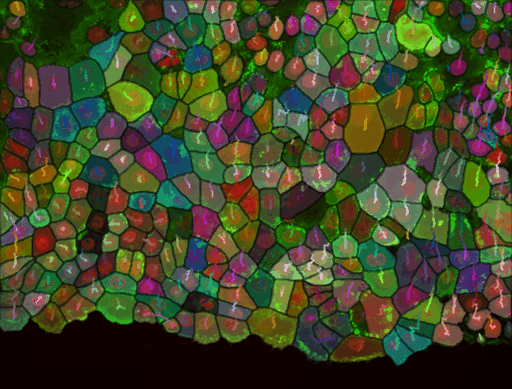
As stated above, we have added the possibility to pass to cellpose all channels or just one. We exemplified below the impact of this setting. This image comes from the lab of Guillaume Jacquemet and was used in doi:10.1038/s41592-022-01507-1
With all channels:
Channel 1 only, in which nuclei are painted in red:
Channel 2 only, in which the cell cytoplasm appears in green:
Channel 3 only, which is empty: