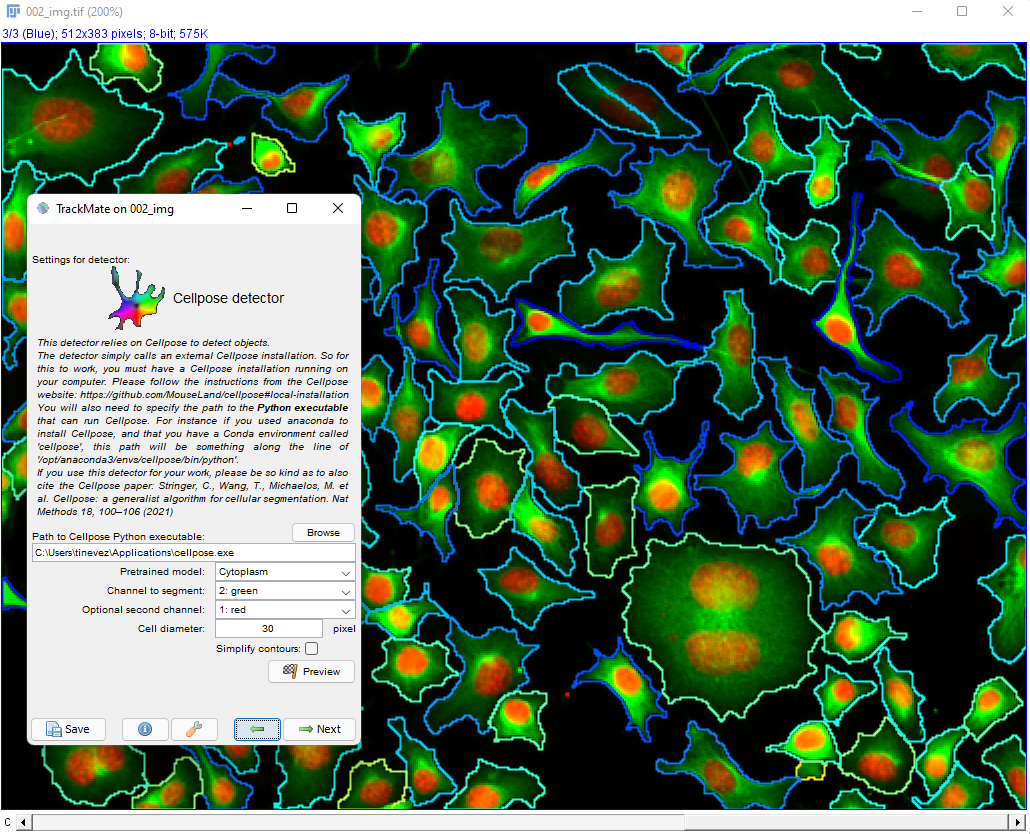
This page describes the advanced version of the cellpose detector in TrackMate, documented here. These two detectors rely both on the cellpose software, but this one offers more configuration options that are hidden in the non-advanced version.
If you use the cellpose TrackMate module for your research, please also cite the cellpose paper:
doi:10.1038/s41592-020-01018-x
Installation
Please see the instructions in the TrackMate-Cellpose documentation page. In particular the Installing cellpose paragraph in the Additional information section.
Usage
Advanced cellpose parameters in the TrackMate UI
We describe here the advanced parameters that are specific to the advanced detector. Core parameters are described in the TrackMate-Cellpose documentation page.
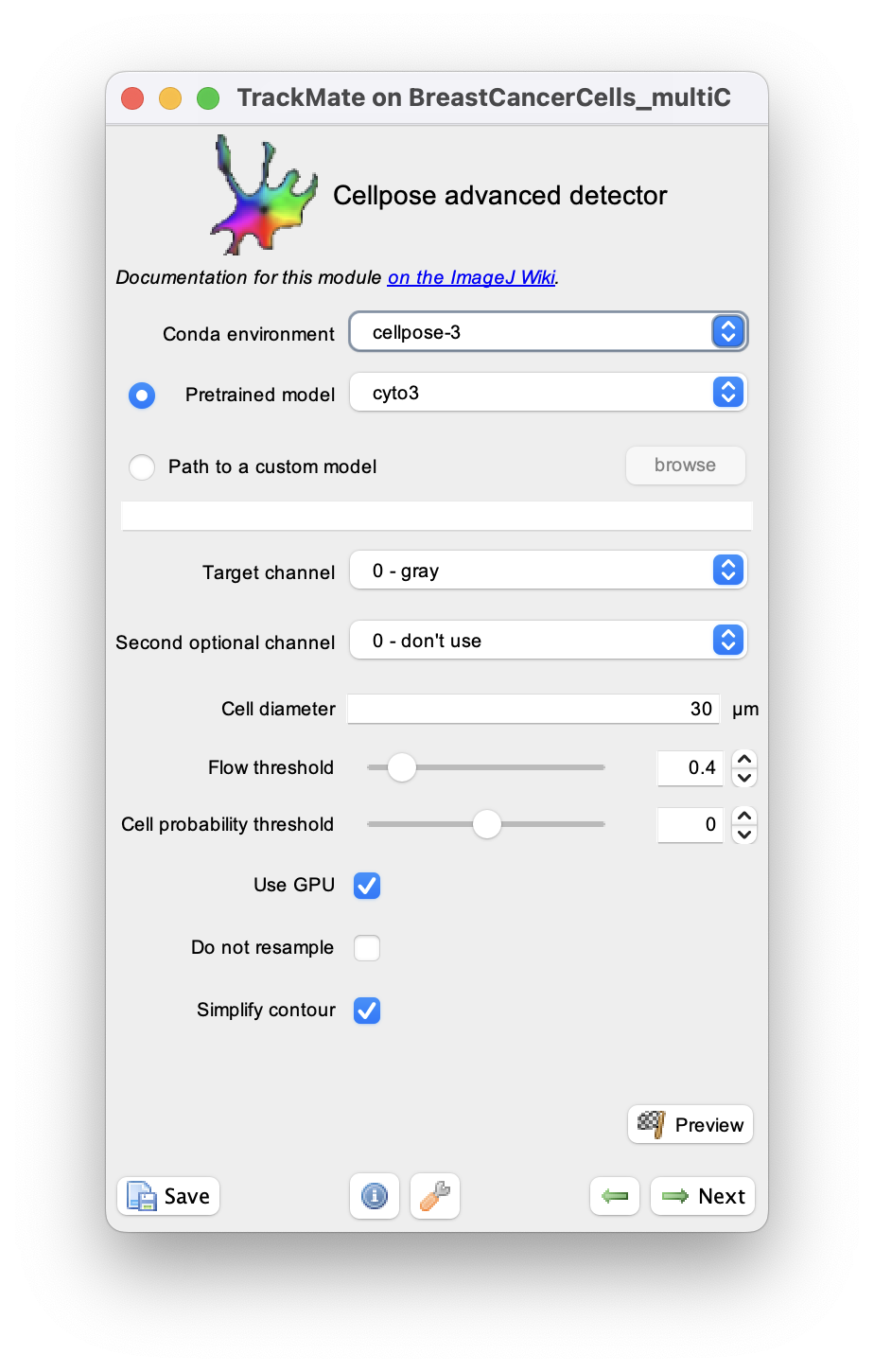
Flow threshold
Cellpose performs an additional step after a first reconstruction of the shape to check the consistency between the calculated cell shapes and the computed flows.
This step estimates the mean squared flow errors based on the cell shapes.
Only flows below the flow threshold parameter will be kept.
This value should be positive. If it’s small, less cells will be kept and with more regular shapes, larger values will give more cells but potentially more ill-shaped.
If it’s 0, this step will not be performed (all detections whatever the shape will be kept).
By default in the non-advanced option, this parameter is 0.4.
Cell probability threshold
One of the neural network output of cellpose is a cell probability map: each pixel contains the value of its probability to belongs to a cell.
This parameter controls the amount of confidence to keep a pixel in the cell: all pixels with a probability above cell_probability_threshold will be kept.
This value can range from -6.0 (more and larger cells) to 6.0 (less and smaller but most likely cells).
By default in the non-advanced option, this parameter is 0.0.
Do not resample
Cellpose resizes your image to have the mean cell size (mean diameter parameter above) equals to the mean diameter of the trained model.
The flows are computed on these rescaled images.
However, the reconstruction of the masks (the cells) can be done either at the same rescaled resolution (resample=False), or at the original image size (resample=True).
It will affect the smoothness of the resulting segmentation and the computing time:
- If your mean diameter > model diameter, the image is downscaled. In this case, resampling will create smoother ROIs but will be slower (the calculation will be done in the original image size, so on larger image).
- If your mean diameter < model diameter, the image is upscaled. In this case, resampling will be faster (running on smaller image), but will find less detections than without resampling.
By default in the non-advanced option, this parameter is set to False.
The images are resampled.