The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
Documentation and latest stable build
Summary
FijiRelax is a generic tool for 3D+t MRI analysis and exploration using multi-echo spin-echo sequences. This work was supported by the French Ministry of Agriculture, France AgriMer, CNIV and IFV, within VITIMAGE and VITIMAGE-2024 projects (program Plan National Dépérissement du Vignoble). This tools is developed in the context of :
- the Vitimage 1 and Vitimage 2024 projects.
- the Aplim flagship project.
Statement of need
FijiRelax is a generic tool for multi-echo spin-echo T1-T2 relaxometry capable of processing a wide variety of MRI images ranging from a plant stem to a human brain (see \autoref{fig:figure1}-b). It has been designed for three types of scientists: i) end-users using a GUI, ii) advanced users able to use a scripting languages to process large number of images, and iii) developers able to adapt and extend the application with new functionalities.
-
End-users using a GUI: this mode is recommended for scientists who are not specialists in image processing nor programming. Download FijiRelax through the official Fiji release, and follow the step-by-step installation instructions, as well as the hands-on tutorials built on the test dataset hosted at Zenodo [@fijirelaxDataset]. Then, use the graphical user interface to import and process your own Bruker/NIFTI/Custom data, explore the relaxation maps in space and time using the graphical relaxation curve explorer and export your results as 2D/3D/4D TIFF images. This mode is also recommended for studying new datasets or new biological questions. Among the interface features, the plugin provides a graphical explorer to visualize the relaxation curves, and the estimated PD-weighted T1 and T2 distributions over customizable areas of interest. In 5D hypermaps, the distributions at each time-point can be displayed simultaneously, giving access to valuable information on water distribution in tissues and its evolution during the monitoring period.
-
Advanced users: this mode can be used by scientists with programming skills. Load the sample BeanShell scripts provided in repository in https://github.com/Rocsg/FijiRelax/tree/-/test/Scripts . Select a script by dragging it into the Fiji interface and run the scripts to reproduce the results shown in the paper in figure1: import a dataset, convert it to an HyperMap (see figure1-e), compute the parameter maps. Then, adapt these scripts to your needs, including processing your own data and batch-processing multiple experiments.
-
Developers: this mode is for programmers fluent with Java and Maven. Start by exploring the FijiRelax API: API Overview. Build your own tools on top of the FijiRelax library, provided as a jar file hosted at maven central repository (Artifact), by indicating FijiRelax as a dependency in your POM file and run the unit tests. FijiRelax is hosted on a github public repository (https://github.com/rocsg/fijirelax) and developers can offer to contribute to its development, and extend it by requesting features, or proposing new features.
Developer documentation:
Plugin features
- Proton density, T1 and T2 maps computation from multi-echo spin-echo sequences (multiple TR and/or TE)
- Parameters estimation by fitting noise-corrected mono- and biexponential decay models
- Automatic correction of spatial drift and deformations for long T1 or T2 sequences
- Exploration of T1/T2 distribution in ROI over time
- Operable through a GUI, or scriptable for batch processing of large datasets
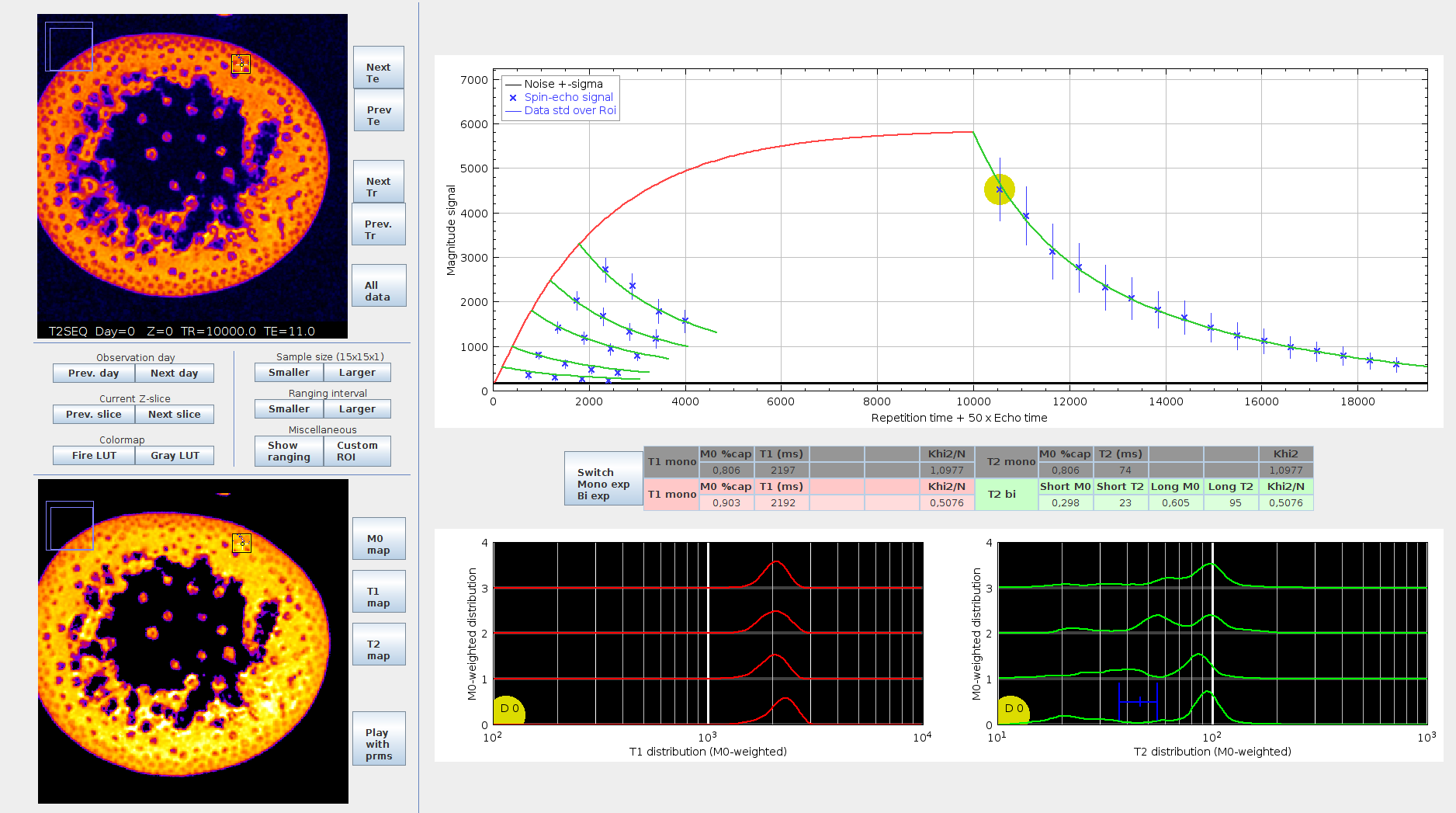
Time-lapse exploration of parameters in a plant under drought stress
Dataset for testing purpose
A comprehensive dataset can be found on Zenodo at https://doi.org/10.5281/zenodo.4518730
Installation
The following video guide you throughout the installation process, and take a first tour of FijiRelax functions. Window users, warning. You have specific operations to run at the end of this section.
In order to install FijiRelax on your computer, please follow these steps:
1. *(if needed) *Download and install Fiji from https://fiji.sc/ ; start Fiji, and let it automatically update. Then restart Fiji.
2. Open Fiji, run the Update manager Help › Update. Click on “OK” to close the first popup windows, then click on the button Manage update sites….
3. In this list, activate ImageJ-ITK by checking the corresponding checkboxes. Don’t close the window, or reopen it if you read this too late.
4. Add the Fijiyama repository (by clicking on the button Add update site, and filling the fields : name = “/plugins/fijiyama”, site = https://sites.imagej.net/Fijiyama), then check the associated checkbox. Now you can click on Close and apply the modifications.
5. Restart Fiji: a new FijiRelax entry should be available in the menu Plugins › Analyze. If not, go back to the Update Manager, and check that the repositories ImageJ-ITK and Fijiyama are correctly selected.
6. If you are a Windows user: there is specific issues between an external package ImageJ-ITK and the current JDK shipped with ImageJ. Whether or not you understand this point, please follow the documented procedure to escape this. You have to use ImageJ with a different JAVA version. It is well explained there: https://imagej.net/learn/faq#on-windows . You can find for example an OpenJDK 8 at OpenLogic, then install it, and when asked, check the box to select that it have to build a JAVA_HOME variable (replace a red cross symbole by a hard-disk symbol when asked, you’ll see it). After that you remove the java or jre dir in your Fiji.app, as said, and it’s ok!
Preparing your data
FijiRleax needs properly formatted dataset:
- Nifti 4D images, or a set of Nifti 3D images
- Dicom dirs with 3D images, or a set of dirs with 2D images
The interface
FijiRelax interface have four main panels :
- With the first panel, you can import / open / export data.
- The second panel holds the processing routines.
- The third panel contains the explorer button.
- The fourth panel has additional helper functions.
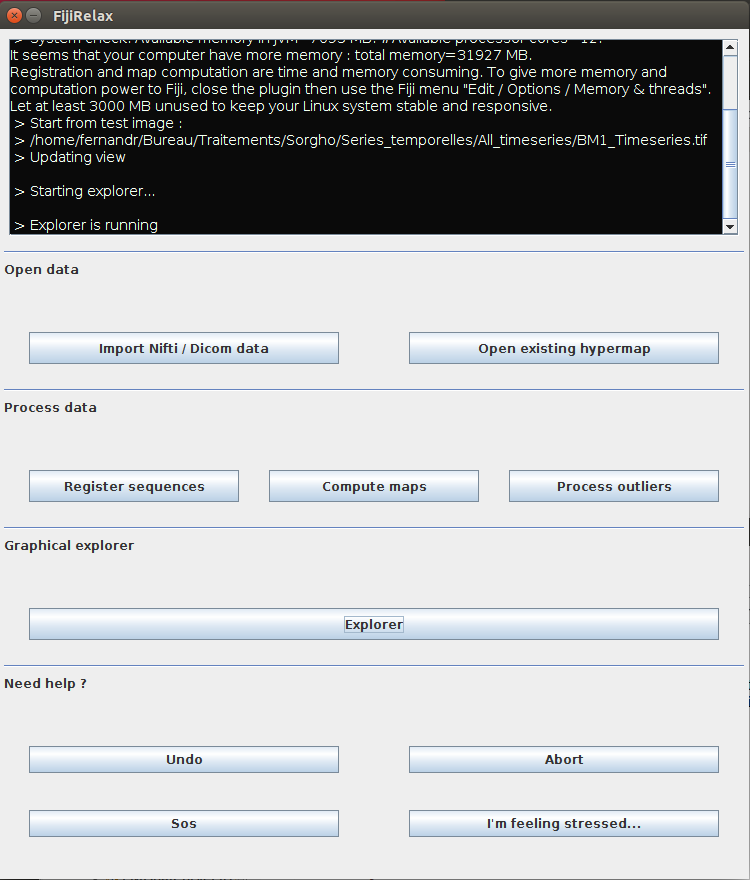
FijiRelax main window
Tutorials
Tutorial part 1: proton density, T1 and T2 time-series from 3D dicom data of a sorgho plant
Tutorial part 2: from 4D HyperMaps to time-lapse plant physiology monitoring
HyperMap data structure
The output image is a 4D MR hyperimage. The “channels” slicer helps you to explore the 4th dimension, that is the images computed, and the input spin echo images. In detail :
- Channels 1,2 3 are respectively the M0 map, T1 map, T2 map (see this information in the slice title, just upside the image pixels)
- Channels 4,5, ….. NR-3 are the successive NR repetition times of the “T1 sequence”, in increasing order.
- Channels NR-2,….. NR-2+NE are the successive NE echo times of the “T2 sequence”, in increasing order.
Unit for the channels 2 and 3 are milliseconds, what mean you can use it like it, without any additional conversion.
For time-lapse experiments, one can compute such a 4D MR hyperimage at successive timepoints, and register and combine them in a 5D MR hyperimage (the same, with an additional slicer to walk through time). Registration and data combining can be done using the series registration mode of the Fijiyama plugin.
The science behind
This plugin compute M0, T1 and T2 maps pixelwise from a given set of spin-echo sequences, acquired with different repetition times and/or different echo times.
First a 3d registration is computed to align precisely the successive images, using libraries of the Fijiyama plugin. Then the rice noise level is estimated, and the M0, T1 and T2 parameters are estimated, fitting mono or bi-exponential curves, corrected with the measured rice noise. For more information, see the paper in next section.
Citing this work
- Romain Fernandez, Cédric Moisy, Christophe Goze-Bac, Maïda Cardoso, Rahima Sidi-Boulenouar, Jean-Luc Verdeil, 2021 «FijiRelax: Fast and noise-corrected estimation of MRI relaxation maps in 3D + t» under review
Software dependencies acknowledgements
- Johannes Schindelin et al for Fiji (Schindelin et al., 2012)
- Karl Schmidt for MRI Analysis Calculator and CurveFitters
License
This program is an open-source free software: it can be redistributed and/or modified under the terms of the GNU General Public License as published by the Free Software Foundation (http://www.gnu.org/licenses/gpl.txt).
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
