The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
Current Fijiyama version : Handsome Honeysuckle (last release : October 16th, 2022).
The plugin Fijiyama (Yet Another Matching and Alignment tool for Fiji) is a generic tool for registration and alignment of 3D image series collected from various imaging modalities (MRI, X-rays, Microscopy, Photography, …).
This work was supported by the French Ministry of Agriculture, France AgriMer, CNIV and IFV, within VITIMAGE and VITIMAGE-2024 projects (program Plan National Dépérissement du Vignoble). This tools is developed in the context of :
- the Vitimage 1 and Vitimage 2024 projects.
- the Aplim flagship project.
Multi-modal time-series registration
Context: 3D images acquired with various imaging devices or during monitoring experiments often come with different orientations and positions, making it difficult to analyze. Fixing this misalignment in 2D is difficult and become intractable in 3D. In addition, as each modality highlights different structures, images from multiple devices can be very dissimilar. In this situation, structures and phenomenon analysis can be compromised.
Goal: Fijiyama aims to address these issues by helping users to register, align and combine multi-modal 3D images or time-series in a unique reference geometry. After registration, stacks are combined in a coherent multi-modal and/or multi-time volume that can be explored using the ImageJ viewer.
Versatility: Fijiyama is a generic tool, efficient at any scale, tissue or imaging modality. It has been tested on the most common imaging approaches in biology and medicine: MRI, X-rays, photography…
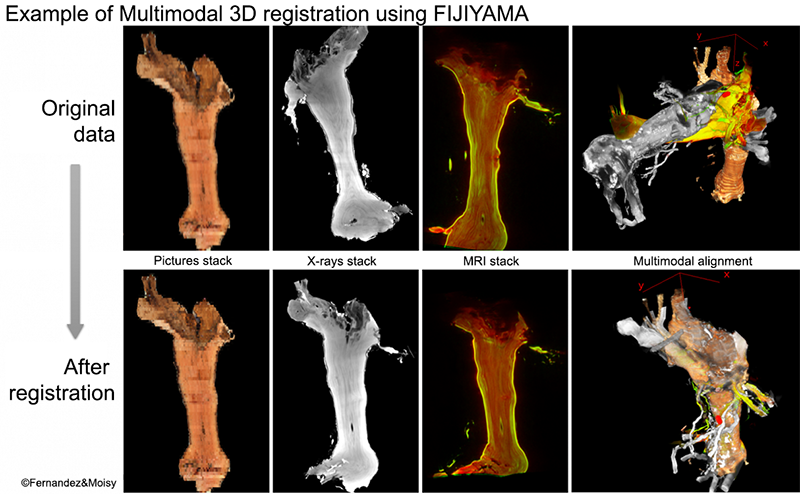
Dataset with DOI for testing purpose
A comprehensive dataset can be found at this link on dryad.
Demo
The following video shows an example of Fijiyama application: the registration of two stacks of MRI 3D images, collected on a grapevine cutting at different dates(see the “Training tutorial” for details).
Plugin features
- Two images registration (training / settings mode)
- N-times / M-modalities 3D series registration
- Composition of successive ITK transformations into a single transformation
- Transformation of a 3D image (or a 4D / 5D hyperimage) with an ITK transformation
In order to address the complex task of multi-modal registration, computation is based on the Block Matching algorithm (Ourselin et al., 2000), a generic purpose registration algorithm, developed in the field of medical imaging. Algorithms from ITK and Simple ITK (Elastix algorithms) are available in the plugin as an option. To learn more about the algorithmic bases of the plugin, see section “The maths behind”.
Installation
In order to install Fijiyama on your computer, please follow these simple steps: Window users, warning. You have specific operations to run at the end of this section.
1. (if needed) Download and install Fiji from https://fiji.sc/ ; start Fiji, and let it automatically update. Then restart Fiji.
2. Open Fiji, run the Update manager Help › Update. Click on “OK” to close the first popup windows, then click on the button Manage update sites….
3. In this list, activate ImageJ-ITK by checking the corresponding checkboxes. Do the same for IJPB-plugins
and add the Fijiyama repository (by clicking on the button Add update site, and filling the fields : name = “/plugins/fijiyama”, site = https://sites.imagej.net/Fijiyama), then check the associated checkbox.
4. Restart Fiji: a new Fijiyama entry should be available in the menu Plugins › Registration. If not, go back to the Update Manager, and check that the repositories ImageJ-ITK, and Fijiyama are correctly selected.
5. For Windows users, there is a compatibility problem with the JAVA package supplied with ImageJ, so you must install a different version of JAVA. For more information on this issue, see doc at https://imagej.net/learn/faq#on-windows. To solve the problem:
- a) Download and install the latest version (.msi installer file) of OpenJDK 8 from the OpenLogic website (https://www.openlogic.com/openjdk-downloads).
- b) During installation, click on the red cross in front of the “Define JAVA_HOME variable” option and select the “Install on local disk” option (it changes the red cross to a hard disk icon).
- c) Once installation is complete, go to the Java folder of your Fiji/ImageJ software (usually …/fiji-win64/Fiji.app/java/) and delete the contents of the directory. If no bundled Java is found, ImageJ will fall back to your “system Java”, which is indicated by the JAVA_HOME environment variable you’ve just installed.
- d) Restart ImageJ and enjoy FIJIYAMA, it should work!
The following video shows a tutorial for Fijiyama installation:
Preparing your data
In order to register your data, Fijiyama needs properly formatted dataset:
- For two images pairwise registration : Two 3D stacks/images of the same object. Use your own data or the provided example datasets here: case study 1, two-images registration
- For series registration : N 3D stacks/images of the same object. Use your own data or the provided example datasets here: case study 4, times series registration.
- An empty “Output” directory, that will be used to store the Fijiyama configuration file, the intermediate results (transformations) and the final results (transformations and resulting 3D images). The configuration file (*.fjm) keep a full track of your work. When saving the current registration experiment, Fijiyama stores the successive registration steps and the associated computed files. When restarting later from this point, you can review the whole process, and “undo” one or more steps.
Before starting registration, check your images calibration:
- **Intensities: ** verify there is a proper contrast between your object of interest and the background when opened in Fiji/ImageJ. If needed, use Image › Adjust › Brightness / Contrast, and set the min and max values, then close the Brightness / Contrast window.
-
**Voxel sizes: ** using ImageJ, check the image properties Image › Properties… and correct the 4 central values, if needed. Unit of length **should have another value than “pixel” ** (it can be “mm”, “cm”, “µm” for example). Check coherence of pixel width / height / depth according to your unit of length. If needed, open the “info” menu (Image Show info…), to get dicom parameters, or any other parameters. To visualize if the voxel depth is coherent with the pixel size, along X and Y axis, you can render your object as a 3D volume using the 3D viewer (plugins 3D viewer).
Once signal intensity and voxel sizes have been checked, save your modified images. Then run Fijiyama, and select the two images registration mode or the series registration mode.
The registration manager window
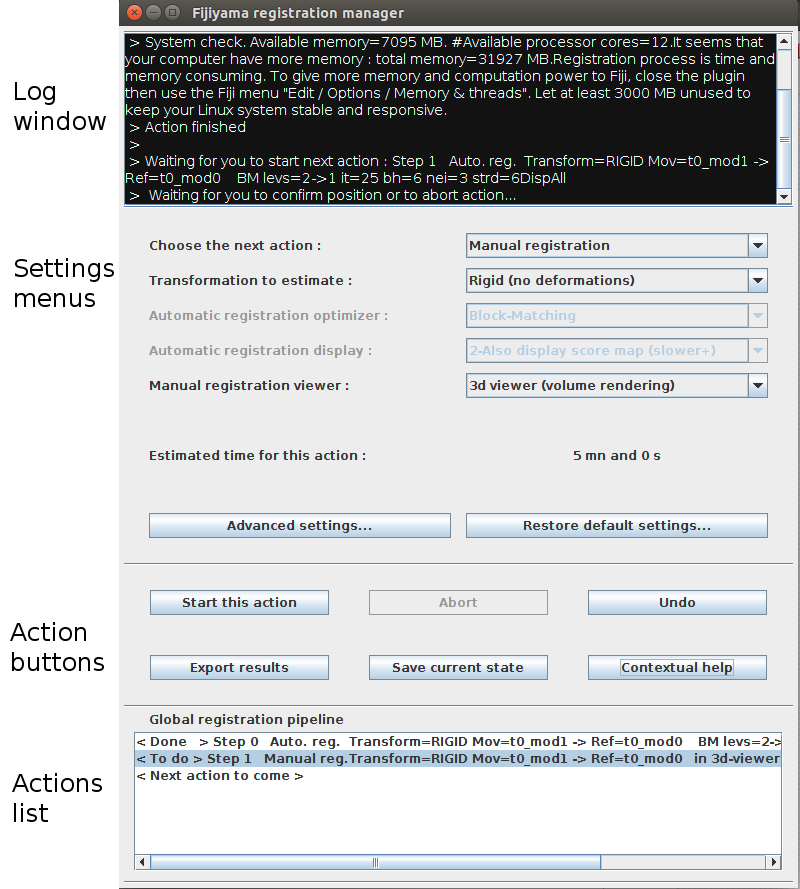 Once a registration scenario starts, you can interact with the registration manager window through (see image) :
Once a registration scenario starts, you can interact with the registration manager window through (see image) :
- The log window : a guide to remember last operations, and to understand what the plugin expects from you
- The settings menus and buttons : useful to set the parameters of the future action
- The action buttons : the first line (Start, Abort, and Undo) drives the lifecycle of the current action, defined by the menus and the current settings. Save can be used to save the progress made in the registration, as a “checkpoint”. Export computes the resulting aligned 3D images, according with the succession of transformations performed. Help can be used at any time to open a contextual help message.
- The actions list : stores the successive steps accomplished during the registration process.
Tutorial 1 : Two images registration
In order to achieve an efficient registration of two 3D images, we recommend the following procedure:
- **Step 1 : ** Prepare your data or download the example set: case study 1, two-images registration
- Step 2 : ** Select **‘Manual registration’ in the first menu and run it. Superimpose roughly the two volumes manually (centers should be roughly aligned, with angle < 15 degrees). Press on ‘Position ok’ (green button) when ready. This first step will help Fijiyama finding the correct orientation.
- Step 3 : ** Select **‘Automatic registration’ with default parameters (Block matching). If you want to monitor the registration during the run, select the ‘Display automatic registration’ in the “Manual registration viewer” menu. Before starting this action, notice the estimated time required to complete the action (calculated from your computer settings).
- Step 3-bis **(optional, depending on the data) : Select **‘Automatic registration’ with default parameters (Block matching), and select “Vector field” in the “Transformation to estimate” menu.
- Step 4 : ** Select **‘Align both images with XYZ’ with default parameters. Turn the scene to improve its orientation, if needed for a more convenient representation of your data (production of figures or analyses). In the example dataset, we choose to align the red cylinder with the white lines (Z axis), and align the wounding point (a hole in the surface) with the axis X or Y.
- Step 5 : ** Save the final transformation using the **‘Save current state’ button.
- Step 6 : ** Export the registered 3D images in the “output/Exported_data” directory previously selected, using the **‘Export results’ button.
Tutorial 2 : Multimodal time-series registration
Once comfortable with the “two images registration” module, try a “series registration” following these steps. If you feel lost, you can rely on the tutorial here :
- **Step 1 : ** Prepare your data or download the example data set : case study 4, times series registration
Data names should respect a generic form, for example : img_t{Time}_mod{ModalityName}.tif , where {Time} represents the successive values for each time point, and {ModalityName} represents the name of each modality. In the example dataset, this is already done, since the image names are : MRI_D0.tif , MRI_D1.tif , MRI_D2.tif , MRI_D3.tif, which corresponds to a monomodal series with the generic expression MRI_D{Time}.tif .
- **Step 2 : ** Use the “Two images” training module to identify the best pipeline of actions for registering your images, from a modality to another, or from an observation time to another.
- Step 3 : ** Start the module **‘Series registration’, and follow the instructions. The manager will ask the pipeline for inter-time registration, then for inter-modalities registration. Once done, it will compose these steps into a full pipeline and process. You can repeatedly press on ‘start this action’, and execute all the steps.
- Step 4 : ** Once all actions needing human intervention are performed, use the **‘Chain run’ button to ask Fijiyama to execute successively all the steps automatically.
- *Step 5 : ** Run the last actions. The manual alignment step will set the reference geometry in the image axis, and the save and export steps will stores the actions and the results. The results are saved in the *output/Exported_data directory.
Complementary features
Compose transformations
Chaining linear and/or dense transformations is one of the plugin core features for series processing. This function can also be used as a helper.
Guessing you already computed :
- the transform T12 that aligned Image1.tif with Image2.tif
- the transform T23 that aligned Image2.tif with Image3.tif
You may want to align Image1 with Image3 in order to compare the three 3D images altogether. However, applying successive transformations will result in a strong blurring effect. In order to avoid this blur, you may want to combine the two transformation steps into a single one.
Use the compose transformations function to run in one go the successive transformations to be composed (in the example T12 then T23), and produce the resulting combined transform.
Apply a transformation to a 3d/4d/5d Image
This module is a helper function to apply an ITK transformation to a 3D image. Since the Gorgeous Grapevine version, this function can be applied to a 4D/5D hyperimage, the transformation being interpreted as a 3D transformation, and applied to each channel / frame of the hyperimage before the transformed 3d images are gathered back in an hyperstack).
Versatility Tests
Fijiyama plugin has been tested on several data (including vegetal and human tissues), at different scales (from micro to macro imaging), and on different imaging modalities (microscopy, MRI, X-rays, and photographies).
See an example in the video below : Human abdomen CT-scan and MRI registration (Courtesy of Dr. Samuel Mérigeaud, Tridilogy SARL) <div class="video"><iframe src="https://www.youtube.com/embed/ETCnWIqoDI0" width="400" height="300" frameborder="0" webkitallowfullscreen mozallowfullscreen allowfullscreen allow="accelerometer; encrypted-media; gyroscope; picture-in-picture"></iframe></div>
Accuracy Test
Fijiyama can be used to quantify the resulting mismatch. This measurement process is integrated in the plugin as an extra functionality, to help users evaluating their results, as illustrated in this video : “Fijiyama : quantifying average mismatch after registration”
Versions of Fijiyama
Fijiyama is a living project, with frequent updates. Thanks to the Fiji updater, your version is updated each time the Fiji update starts.
Major updates include new features released, or major refactoring, while minor updates (change in the release time) should not modify deeply the behaviour of the plugin, to keep results reproducible. These information can be seen in the launching interface of Fijiyama.
Major updates are symbolized with a change in the plugin version name. For minor updates, only the release time is modified.
| Version name | First release | Last release | Integration of new features | Modifications causing reproducibility issues |
| Elastic Eucalyptus | 2020/02/10 | 2020/02/20 | Integration of the multimodal series registration feature. Redefinition of default settings in the settings editor. | Default settings changes in the settings editor |
| Felicity Ficus | 2020/02/20 | 2020/03/02 | Integration of the time-lapse multimodal series feature. Versioning of serialized files | 1th march: versioning of serialized files starts. Previously serialized files are then open with default values for level min, level max and block matching iterations. |
| Gorgeous grapevine | 2020/03/20 | 2020/03/20 | Hyperimage support (registration of 4D / 5D multi-channels or multi-time images | |
| Handsome Honeysuckle (Future release) | Mid-june 2020 | … | New transformation models : Rigid 2D, Similarity 2D, Dense 2D. Support of extensive grandeurs (images which intensity have to raise when it is shrinked). Support of hyperimage with varying number of channels | … |
Possible issues and recommendations
-
Dealing with large datasets: biological images are sometimes very large. In the field of time-lapse microscopy, BigStitcher plugin handles such data, using BigDataViewer, as its registration algorithm, perfect for monomodal data of lightsheet microscopy, is well fitted to this task.
Fijiyama is based on the Blockmatching registration algorithm, robust for inter-modality. This algorithm is memory consuming and computation intensive. When starting Fijiyama, the plugin analyzes your computer settings and might propose to subsample your dataset to fasten the registration process. This will not impact the appearance of the final results since the exportation procedure is performed using the initial images, at the chosen resolution.
- Translation in 3D under Windows: under Windows, translation mode in the 3D_Viewer using SHIFT+drag can be hazardous. To address this, we associated surnumerous controls to the numerical keypad. Using “4” and “6” keys, you can translate the selected volume along the X axis. Using “8” and “2” along the Y axis. Using “5” and “0”, along the Z axis. Rotations can be handled with “7” and “9” (X axis), “1” and “3” (Y axis), and characters “p” and “o” for the Z axis.
- 3D view disappears: depending on the sequence of translations / rotations applied to the scene, the 3D viewer of Fiji/ImageJ sometimes “freezes”. To solve this problem, turn the objects to a 90 degrees’ angle, and the viewer should come back.
-
Other issues? please tell us ! Try to “repeat” your bug from the beginning, identify the context producing the bug, then send an email to corresponding author, including :
- a compressed copy of your output directory
- the copy of the content of the ImageJ log window (probably a hundred lines of sentences in red telling where Exceptions were encountered)
- Any useful commentary
- if possible, the content of the black log window
- if possible, a link to the dataset used
The maths behind
Registration of two images is achieved estimating a geometrical transformation that can be applied to an image (the “moving one”), to superimpose it with another image (the “reference” one).
Transformations
Fijiyama handles the following transformation families :
- Rigid transformations (translation + rotation), estimated using VIB-lib
- Similarities (rigid + isotropic homothetic factor) estimated using VIB-lib
- Dense vector field (a generic representation of a non-linear transformation), estimated using Gaussian interpolation
For more details about transformation, check the supplementary data of the official publication (see section “Cite this work” below)
Optimizers
Common categories of registration optimizers, associated with different registration strategies :
- ITK (Yoo et al. 2002)’ iconic algorithm’ optimizes iteratively a transformation. The optimization process is guided by the superposition improvement measured using a global similarity measure between the two 3D images. Choosing this measure, we assume a relationship between intensities of reference and moving images, and that this relationship is valid and identical in any point of the image. That weak assumption can lead to robustness issues when dealing with multi-modal registration.
- The Block-Matching algorithm (Ourselin et al. 2000) is a hybrid method, using a similarity measure to establish correspondences between subparts of images, then using these correspondences to compute a global transformation for the whole image. Assuming that the relationship between intensities of reference and moving images can be valid in a local neighborhood, blockmatching uses it to compare subparts of the 3D images to identify correspondences. This algorithm is the default one in Fijiyama.
- Fijiyama does not have a “geometric” registration algorithm yet. Principle of such algorithms is to extract features from each image, and to establish correspondences between extracted features. Such approach can lead to faster but less robust algorithms as it is difficult to define a feature extraction method that is modality-independent, object-independent, and scale-independent. But for monomodal registration, such algorithms can provide great success, like BigStitcher with microscopy images
Limitations
For large datasets (15 and more timepoints) or images (1GB+), specific tools should be used. Here is a non-exhaustive list of plugins for ImageJ and/or Fiji, available for 3D registration of large datasets: BigStitcher, Atlas_Toolkit, TrakEM2 or Elastix wrapping plugin. Outside from the ImageJ world, for people who are not afraid to script, Elastix is a proper tool for series, and Slicer can also do a great job. Limited to two-images registration, MedInria is a very powerful and popular tool for medical images, which shares a lot of algorithmic basis with Fijiyama.
Citing this work
- R. Fernandez and C. Moisy, (2020) Fijiyama: a registration tool for 3D multimodal time-lapse imaging. Bioinformatics
Hall of fame: the Fijiyama community
The Fijiyama community is growing : we have astronomers, plant scientists, biomedical experts, coming from all over the world. We are open to bug reports and feature requests. If you like Fijiyama, please send us an email to romainfernandez06ATgmail.com !
Here is a short-list of some Fijiyama users who helped us conceiving the software, fixing some bugs, or developing new features. We are grateful to them :
 |
Cedric Moisy, PhD - French Wine and Vine Institute, Montpellier, France. Topic: Detection of tissue degradation and time-lapse monitoring of grapevine trunk diseases |
 |
Katie Villa, PhD - Massachussets Institute of Technology, Nedivi lab, Boston, USA. Topic: Neuronal plasticity |
 |
Esther Wershof, PhD - Sloan Kettering Institute for Cancer Research, New York, USA. Topic: Embryonic organogenesis |
 |
Benjamin Padman, PhD - Monash Biomedicine Discovery Institute, Melbourne, Australia. Topic: Cellular mechanisms of mammalian mitophagy |
 |
Junya Matsuno, Division of Earth and Planetary Sciences, University of Kyoto, Japan. Topic : Extraterrestrial matter analysis from Hayabusa spatial probes. |
Contact
Issues, feature request ? Please contact romainfernandez06ATgmail.com
References
- Hörl, David, Fabio Rojas Rusak, Friedrich Preusser, Paul Tillberg, Nadine Randel, Raghav K. Chhetri, Albert Cardona, et al. 2019. « BigStitcher: Reconstructing High-Resolution Image Datasets of Cleared and Expanded Samples ». Nature Methods 16 (9): 870-74. https://doi.org/10.1038/s41592-019-0501-0.
- Ourselin, S., A. Roche, S. Prima, et N. Ayache. 2000. « Block Matching: A General Framework to Improve Robustness of Rigid Registration of Medical Images ». In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2000, édité par Scott L. Delp, Anthony M. DiGoia, et Branislav Jaramaz, 1935:557-66. Berlin, Heidelberg: Springer Berlin Heidelberg. https://doi.org/10.1007/978-3-540-40899-4_57.
- Schindelin, Johannes, Ignacio Arganda-Carreras, Erwin Frise, Verena Kaynig, Mark Longair, Tobias Pietzsch, Stephan Preibisch, et al. 2012. « Fiji: An Open-Source Platform for Biological-Image Analysis ». Nature Methods 9 (7): 676-82. https://doi.org/10.1038/nmeth.2019.
- Yoo, Terry S., Michael J. Ackerman, William E. Lorensen, Will Schroeder, Vikram Chalana, Stephen Aylward, Dimitris Metaxas, et Ross Whitaker. 2002. « Engineering and Algorithm Design for an Image Processing Api: A Technical Report on ITK–the Insight Toolkit ». Studies in Health Technology and Informatics 85: 586-92.
Software dependencies acknowledgements
- Johannes Schindelin et al for Fiji (Schindelin et al., 2012)
- Johannes Schindelin et al. for the VIB-lib
- Benjamin Schmid et al for the 3D Viewer (Schmid et al., 2010)
- The ITK and SimpleITK teams (Yoo et al., 2002; Lowekamp et al., 2013)
License
This program is an open-source free software: it can be redistributed and/or modified under the terms of the GNU General Public License as published by the Free Software Foundation (http://www.gnu.org/licenses/gpl.txt).
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.

