The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
{| |
style=”vertical-align:top” |<div class="thumbnail" >
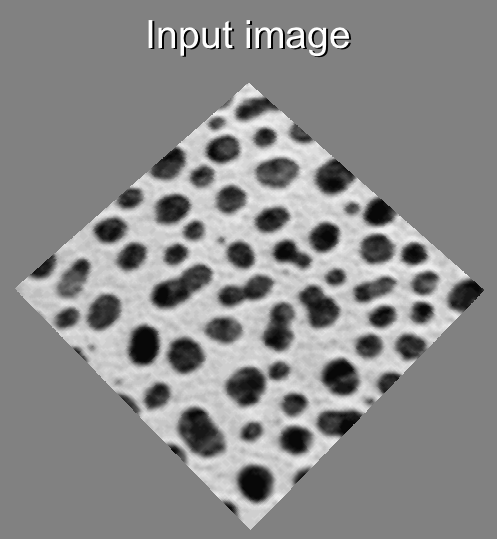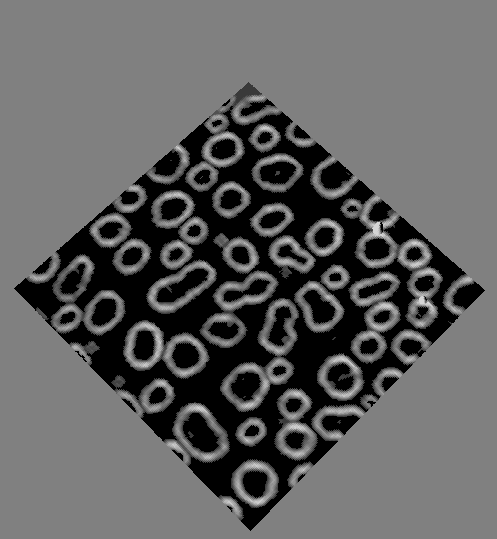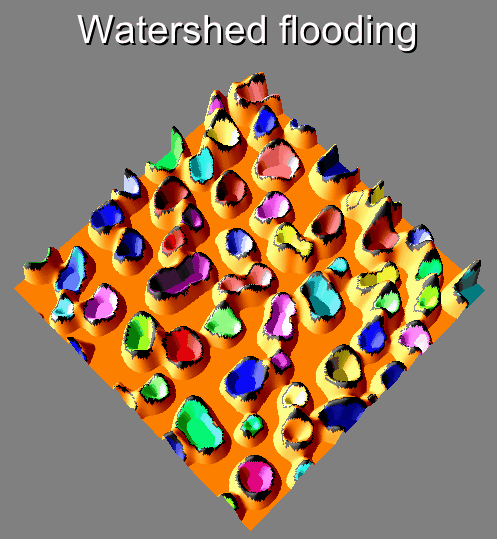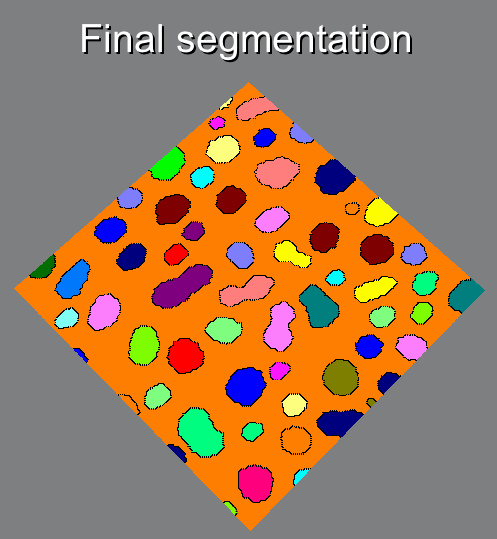 Visual description of the different steps of the Morphological Segmentation plugin.
</div> |}
Visual description of the different steps of the Morphological Segmentation plugin.
</div> |}
Introduction
 Morphological Segmentation is an ImageJ/Fiji plugin that combines morphological operations, such as extended minima and morphological gradient, with watershed flooding algorithms to segment grayscale images of any type (8, 16 and 32-bit) in 2D and 3D.
Morphological Segmentation is an ImageJ/Fiji plugin that combines morphological operations, such as extended minima and morphological gradient, with watershed flooding algorithms to segment grayscale images of any type (8, 16 and 32-bit) in 2D and 3D.
Usage
Morphological Segmentation runs on any open grayscale image, single 2D image or (3D) stack. If no image is open when calling the plugin, an Open dialog will pop up.
The user can pan, zoom in and out, or scroll between slices (if the input image is a stack) in the main canvas as if it were any other ImageJ window. On the left side of the canvas there are three panels of parameters, one for the input image, one with the watershed parameters and one for the output options. All buttons, checkboxes and input panels contain a short explanation of their functionality that is displayed when the cursor lingers over them.
Image pre-processing: some pre-processing is included in the plugin to facilitate the segmentation task. However, other pre-processing may be required depending on the input image. It is up to the user to decide what filtering may be most appropriate upstream.
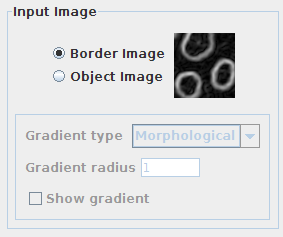
Input Image panel
First, you need to indicate the nature of the input image to process. This is a key parameter since the watershed algorithm is expecting an image where the boundaries of objects present high intensity values (usually as a result of a gradient or edge detection filtering).
You should select:
- Border Image: if your input image has highlighted object boundaries.
- Object Image: if the borders of the objects do not have higher intensity values than the rest of voxels in the image.
When selecting “Object Image”, an additional set of options is enabled to choose the type of gradient and radius (in pixels) to apply to the input image before starting the morphological operations. Finally, a checkbox allows displaying the gradient image instead of the input image in the main canvas of the plugin (only after running the watershed segmentation).
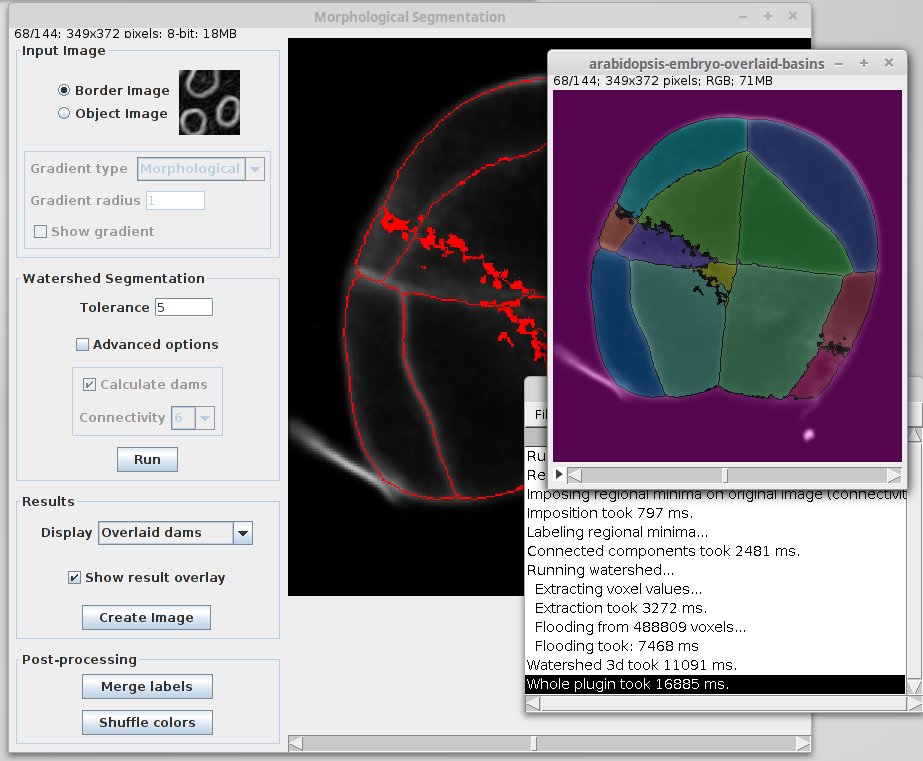
Watershed Segmentation panel
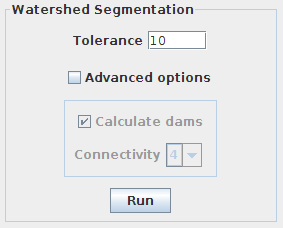 This panel is reserved to the parameters involved in the segmentation pipeline. By default, only the tolerance can be changed. Clicking on “Advanced options” enables the rest of options.
This panel is reserved to the parameters involved in the segmentation pipeline. By default, only the tolerance can be changed. Clicking on “Advanced options” enables the rest of options.
- Tolerance: dynamic of intensity for the search of regional minima (in the extended-minima transform, which is the regional minima of the H-minima transform, value of h). Increasing the tolerance value reduces the number of segments in the final result, while decreasing its value produces more object splits.
Note: since the tolerance is an intensity parameter, it is sensitive to the input image type. A tolerance value of 10 is a good starting point for 8-bit images (with 0-255 intensity range) but it should be drastically increased when using image types with larger intensity ranges. For example to ~2000 when working on a 16-bit image (intensity values between 0 and 65535).
- Calculate dams: un-check this option to produce segmentations without watershed lines.
- Connectivity: voxel connectivity (4-8 in 2D, and 6-26 in 3D). Selecting non-diagonal connectivity (4 or 6) usually provides more rounded objects.
Finally, click on “Run” to launch the segmentation.
If your segmentation is taking too long or you want to stop it for any reason, you can do so by clicking on the same button (which should read “STOP” during that process).
Results panel
 Only enabled after running the segmentation.
Only enabled after running the segmentation.
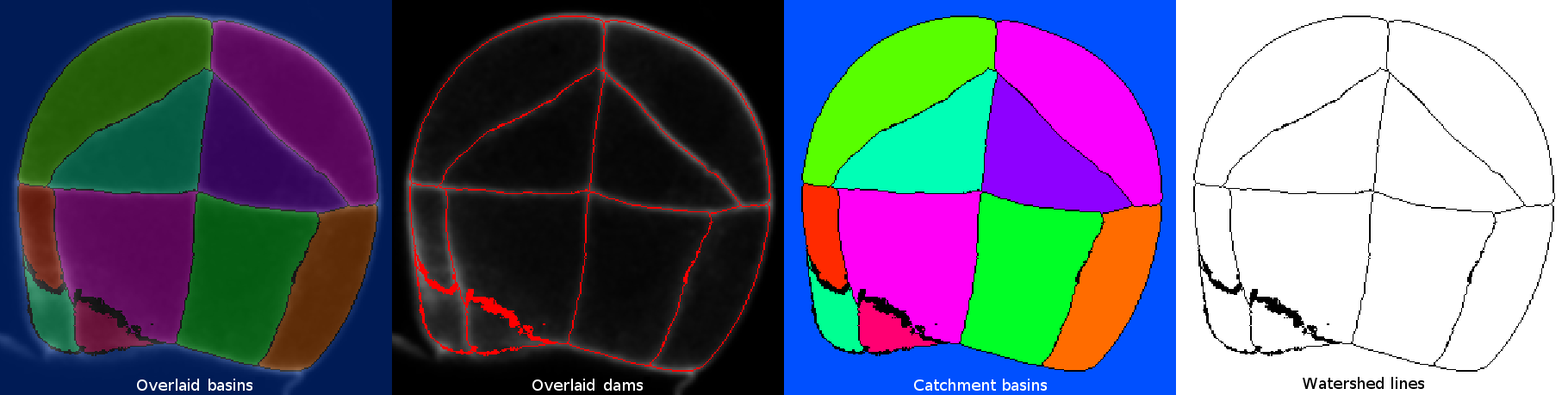
- Display: list of options to display the segmentation results.
- Overlaid basins: colored objects overlaying the input image (with or without dams depending on the selected option in the Watershed Segmentation panel).
- Overlaid dams: overlay the watershed dams in red on top of the input image (only works if “Calculate dams” is checked).
- Catchment basins: colored objects.
- Watershed lines: binary image showing the watershed lines in black and the objects in white (only works if “Calculate dams” is checked).
- Show result overlay: toggle result overlay.
- Create image button: create a new image with the results displayed in the canvas.
Post-processing panel
 Similarly to the Results panel, this panel only gets enabled after running the segmentation pipeline.
Similarly to the Results panel, this panel only gets enabled after running the segmentation pipeline.
- Merge labels: merge together labels selected by either the “freehand” selection tool (on a single slice) or the point tool (on single or multiple slices). The zero-value label belongs to the watershed dams, therefore it will ignored in case of being selected. The first selected label value will be assigned to the rest of selected labels, which will share its color.
Note: to select labels on different slices, use the point selection tool and keep the SHIFT key pressed each time you click on a new label.
- Shuffle colors: randomly re-assign colors to the labels. This is a very handy option whenever two adjacent labels present a similar color.
Video tutorials
Quick start guide
This introductory screencast (which needs audio) explains the basic use of the plugin:
Segmentation of a 3D image
This video tutorial (with audio as well) shows how to use the plugin segment a 3D image (confocal image of an Arabidopsis thaliana embryo, courtesy of Jean-Christophe Palauqui (INRA-Versailles)):
Macro language compatibility
Morphological Segmentation is completely compatible with the popular ImageJ macro language. Each of the buttons in the GUI are macro-recordable and their commands can be reproduced later from a simple macro file.
The complete list of commands is as follows:
- Start the plugin:
run("Morphological Segmentation");
- Select input image:
// select as object image
call("inra.ijpb.plugins.MorphologicalSegmentation.setInputImageType", "object");
// select as border image
call("inra.ijpb.plugins.MorphologicalSegmentation.setInputImageType", "border");
- Run segmentation with specific options:
call("inra.ijpb.plugins.MorphologicalSegmentation.segment", "tolerance=10",
"calculateDams=true", "connectivity=6");
- Toggle result overlay:
call("inra.ijpb.plugins.MorphologicalSegmentation.toggleOverlay");
- Set option to display gradient image:
call("inra.ijpb.plugins.MorphologicalSegmentation.setShowGradient", "true");
- Select display format:
// Overlaid basins
call("inra.ijpb.plugins.MorphologicalSegmentation.setDisplayFormat", "Overlaid basins");
// Overlaid dams
call("inra.ijpb.plugins.MorphologicalSegmentation.setDisplayFormat", "Overlaid dams");
// Catchment basins
call("inra.ijpb.plugins.MorphologicalSegmentation.setDisplayFormat", "Catchment basins");
// Watershed lines
call("inra.ijpb.plugins.MorphologicalSegmentation.setDisplayFormat", "Watershed lines");
- Create new image with the current result:
call("inra.ijpb.plugins.MorphologicalSegmentation.createResultImage");
Complete macro example:
// load the Blobs sample image
run("Blobs (25K)");
// run the plugin
run("Morphological Segmentation");
// wait for the plugin to load
wait(1000);
// select input image as "object"
call("inra.ijpb.plugins.MorphologicalSegmentation.setInputImageType", "object");
// set gradient radius as 1
call("inra.ijpb.plugins.MorphologicalSegmentation.setGradientRadius", "1");
// run segmentation with tolerance 32, calculating the watershed dams,
// 4-connectivity
call("inra.ijpb.plugins.MorphologicalSegmentation.segment", "tolerance=32",
"calculateDams=true", "connectivity=4");
// display the overlaid dams
call("inra.ijpb.plugins.MorphologicalSegmentation.setDisplayFormat", "Overlaid dams");
Installation
Morphological Segmentation is part of the MorphoLibJ library. To install it, you just need to add the IJPB-plugins update site:
1) Select Help › Update… from the Fiji menu to start the updater.
2) Click on Manage update sites. This brings up a dialog where you can activate additional update sites.
3) Activate the IJPB-plugins update site and close the dialog. Now you should see an additional jar file for download.
4) Click Apply changes and restart Fiji.
You should now find the plugin under the sub-menu Plugins › MorphoLibJ › Segmentation.
Note: Morphological Segmentation is only one of the plugins included in the MorphoLibJ library. By following these installation steps, you will be installing as well the rest of plugins in the suite.
Examples
|
style="vertical-align:top" | |
Citation
If you need to cite the plugin, please do so by citing the following paper:
doi:10.1093/bioinformatics/btw413
To cite the implementation, MorphoLibJ’s code repository has its own DOI.
References
- Meyer, Fernand, and Serge Beucher. “Morphological segmentation.” Journal of visual communication and image representation 1.1 (1990): 21-46.
- Soille, P., “Morphological Image Analysis: Principles and Applications”, Springer-Verlag, 1999, pp. 170-171.
See also
- Marker-controlled Watershed, a plugin to perform watershed by flooding from specific seed points or markers.
- Classic Watershed, plugin implementing the original watershed algorithm to segment 2D/3D grayscale images.
- Serge Beucher’s site, with graphic descriptions and animations of the watershed algorithms.
- G. Bertrand’s Topological Watershed site, with papers, lecture slides and source code.
License
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation (http://www.gnu.org/licenses/gpl.txt).
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.