The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
3-way viewer for Block-Face EM image volumes based in ImgLib cells
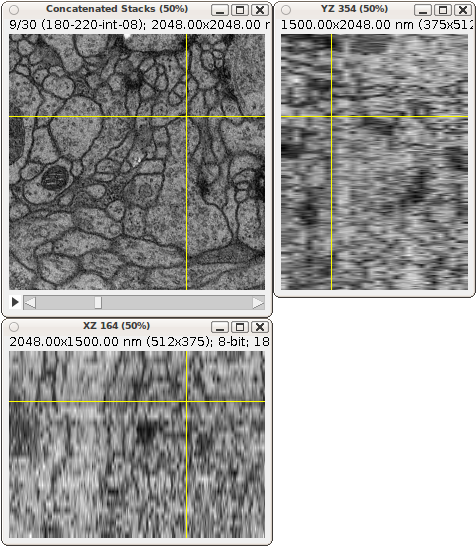 Example of a 3-way view of an anisotropic EM stack. An isotropic stack would show images with the same resolution in the side views.
Example of a 3-way view of an anisotropic EM stack. An isotropic stack would show images with the same resolution in the side views.
Volumetric or higher dimensional data as generated in todays scientific experiments is often too large to be kept in memory entirely. Still, the data needs to be displayed rapidly in multiple views. A typical example is data from Block-Face Serial Scanning Electron Microscopy, displayed through a technique now commercialized as “Gatan 3-way view EM.” See this page for theory and for pictures and examples. Our generic image processing library Imglib is designed to efficiently generate such virtual views from arbitrary n-dimensional image data containers. Still missing is a data back-end that stores the data on the harddisk and provides rapid access to parts of it at multiple scales. The aim of this project is to create such a data back-end and utilize it for an interactive viewer application.
Goal: create a multi-scale disk-stored image data container for Imglib and use it for a rapid multi-view/multi-scale display of very large n-dimensional image data.
Language: any supported by Fiji, preferably Java.
Mentor: Albert Cardona, Mark Longair
Applying machine learning to the image segmentation problem
The term image segmentation describes the task where objects in an image are to be outlined, so that every pixel is connected to either a named object, or background.
Segmentation is traditionally a very difficult problem, especially in the presence of variable lighting, noise, or low contrast.
Many segmentation algorithms have been implemented in Fiji to perform image segmentation, such as Auto Threshold and Auto Local Threshold, but in practice, none of them might work, as they were designed with specific images in mind, and these expectations might not be met by your images.
Recently, a new class of segmentation algorithms has been emerging: segmentation by example. These algorithms require a set of examples from which a model is calculated which can be applied to other – similar-looking – images.
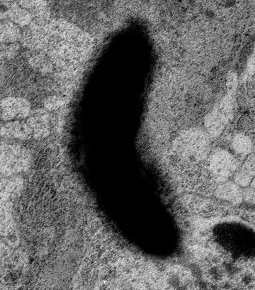
 Segmented neural tissue, painfully done by hand. See the Public data sets
Segmented neural tissue, painfully done by hand. See the Public data sets
We will consider applications for implementations that are either as generic as possible (i.e. they apply to any images), or that try to solve a very specific problem (such as segmenting neurons in serial sections imaged with electron microscopy, or with confocal imaging.)
We have several data sets of images and their corresponding manual segmentations (for training the algorithm). See for example:
- Drosophila larva brain imaged with ssTEM: http://t2.ini.uzh.ch/data.html
- Drosophila embryonic nuclei imaged with confocal microscopy.
You are welcome to use any scientifically-relevant dataset of your choice, but we will give priority to biologically-oriented data sets.
A plugin already exists for Fiji: Trainable Segmentation.
Goal: Implement a number of segmentation algorithms based on machine learning.
Language: Java.
Mentor: Ignacio Arganda-Carreras, Albert Cardona, Mark Longair
Implementing algorithms for Imglib
The new imglib supports dimension-, storage- and data type independent image processing. This library has some algorithms built-in already but there is a strong need to generically implement more general image processing algorithms, storage strategies and data types such as:
- Interpolation (Cubic, Spline, …)
- Entropy Filter, Average Filter, Percentile(Min, Median, Max) Filter, …
- Memory Management for partial image loading
- Color Spaces and Color Space Conversions
- Efficient representation of non-raster images (based on 2d polygonal shapes and 3d/4d meshes)
Goal: Implement generic algorithms for image processing.
Language: Java.
Mentor: Stephan Preibisch, Stephan Saalfeld, Tobias Pietzsch, Albert Cardona
Reparing images with missing data by using contextual information
Implement a simple inpainting method (i.e. restore missing/unwanted parts of the image marked by a ROI) using wavelets: apply the wavelet transform, and then, on each level, use a diffusion algorithm to deduce a smooth signal from the surrounding parts, and finally inverse-transform the wavelet to get the restored image.
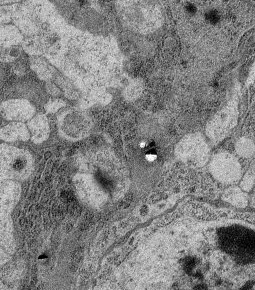
Other approaches are also welcome, such as using information from adjacent serial sections in electron microscopical image volumes. See for example the large black blob at top left which could be restored with information from the next and previous sections.
Goal: implement a plugin for repairing images.
Language: Java.
Mentor: Albert Cardona
Robust blob segmentation
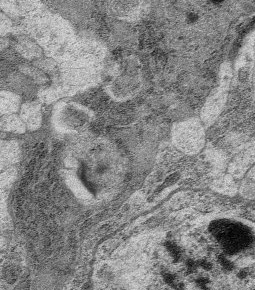
In life sciences, you often cope with round structures of interest. Such round structures can be cells, vesicles, nuclei or similarly shaped objects. While an ellipse might be a good initial fit, the final outline most certainly is not.
It is important to keep in mind that the objects are clearly convex, as they sometimes overlap, and we still want to find the objects correctly. See the adjacent image for an example.
The purpose of this project is to segment in a fully automatic way round, convex structures in biological images. This could be done by using a simple template matching approach for the initial stage, or a Gaussian fit, followed by a fit of the whole outline under the desired constraints.
Goal: Provide a robust blob segmentation algorithm that can work in 2D, 3D and 4D.
Language: Java
Mentor: Albert Cardona