![]()
![]()
This product is a testament to our expertise at KapoorLabs, where we specialize in creating cutting-edge solutions. We offer bespoke pipeline development services, transforming your developmental biology questions into publishable figures with our advanced computer vision and AI tools. Leverage our expertise and resources to achieve end-to-end solutions that make your research stand out.
Note: The tools and pipelines showcased here represent only a fraction of what we can achieve. For tailored and comprehensive solutions beyond what was done in the referenced publication, engage with us directly. Our team is ready to provide the expertise and custom development you need to take your research to the next level. Visit us at KapoorLabs.
This page describes a track corrector module for TrackMate that relies on oneat. It is not included in the core of TrackMate and must be installed via its own update site called TrackMate-Oneat.
The figures and tables mentioned here can be found in our associated publication
Introduction
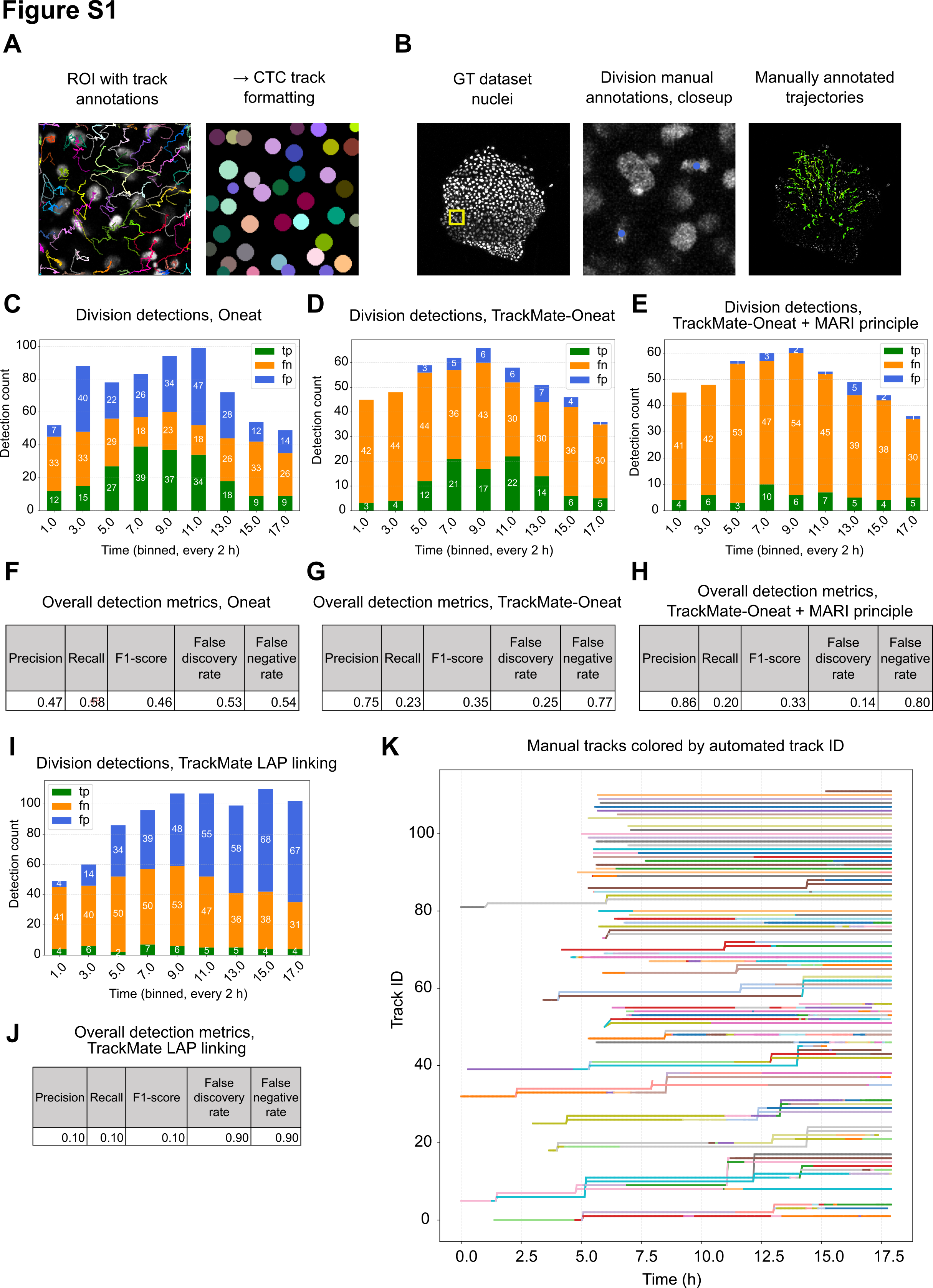
Oneat identifies cell division events as spatiotemporal (TXYZ) coordinates. These coordinates were then used to impose trajectory splitting within TrackMate tracks through our custom-developed TrackMate-Oneat plugin. The plugin optionally applies the MARI (Mitosis Angular Region of Interest) principle, which filters division events to retain only those in which daughter cells emerge perpendicular to the mother cell’s nuclear major axis.
Oneat integration significantly improved mitosis detection compared to TrackMate’s native linking algorithm (TrackMate native branching accuracy = 0.122, TrackMate-Oneat branching correctness = 0.328). By combining Oneat-predicted division locations with trajectory continuity, the system generated more biologically realistic branching structures and reduced the number of false positives typically produced by Oneat alone. However, while applying the MARI principle nearly eliminated false positives, it also led to a reduction in true positive detections (Figure S1E). As such, the user should choose the division detection strategy – Oneat alone or Oneat with MARI filtering – based on the specific goals and tolerance for false positives in their downstream analyses.
Division detection with TrackMate-Oneat
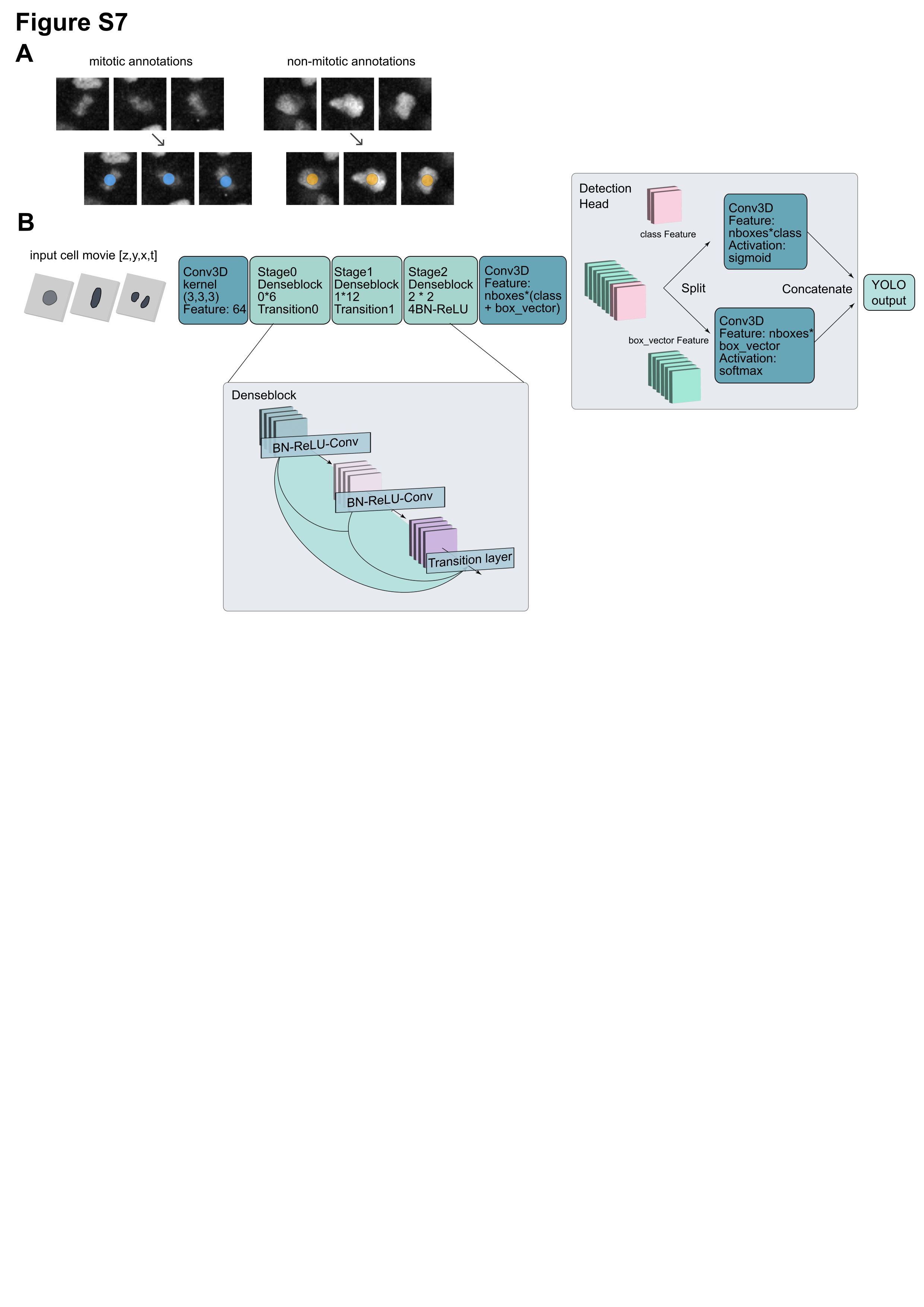
We developed a pipeline leveraging deep learning-based action classification to detect cell division events and use these predictions to improve nucleus tracking. A key component of this system is the Oneat classifier, which was trained to distinguish mitotic from non-mitotic cells based on short temporal sequences of image data. For training, the 3D+t nucleus channel of a dataset was manually annotated for division events. Specifically, mitotic events are annotated on Napari by clicking on the center of the dividing nucleus in a microscopy time series. For each annotated division, a 64 x 64 x 8 voxel crop of 3 time frames was extracted around the clicked location, centered both spatially and temporally on the mitotic event. This creates a positive training sample. To create negative (non-mitotic) samples, a corresponding number of randomly selected locations are extracted from non-dividing nuclei. These negative and positive samples are used to train the model in a supervised fashion, optimizing a binary classification loss to distinguish mitosis from non-mitosis using a DenseNet-based architecture.
For the prediction of division events from whole datasets, the trained Oneat model processes each object identified from a pre-generated nucleus segmentation. For each segmented nucleus at every time point, a temporal window is constructed by cropping 64 x 64 x 8 voxels for 3 frames centered on the nucleus XYZ centroid. These volumes are passed through the Oneat model, which classifies each central frame as either mitotic or non-mitotic.
To combine division events with tracking data of a dataset, the TZYX coordinates of predicted mitotic nuclei were recorded in a CSV file, which was used as input for the TrackMate-Oneat extension of TrackMate. This step uses the predicted division locations to impose a branching point on a trajectory. Suitable daughter cells for mitotic cells are searched from a 16.5 µm search radius from the mother cell, and linking is optimized using a Jaqaman linker algorithm. This biologically-informed relinking improves the completeness and accuracy of lineage tracking, especially in datasets with frequent cell divisions.
To avoid spurious links and ensure geometric plausibility, the pipeline also incorporates the Mitosis Angular Region of Interest (MARI) principle. This constraint limits the search for daughter cells to a angular region from the mother cell’s nucleus principal axis of a fit ellipsoid. Candidate daughter spots were defined as those within a radial distance of the mother spot whose displacement vector , with the candidate spot position, formed an unsigned angle with the mother’s principal axis not exceeding a threshold set by the user. By restricting candidate daughters to fall within a defined angular region of interest, this method eliminates improbable pairings and enhances the biological realism of the reconstructed lineages. This constraint is especially important in dense tissues, where purely distance-based linking may result in incorrect associations.
Explaining the figures
Quality metrics for tracking and division detection. A-B. Division metrics in C-J are based on a dataset where each cell division is manually annotated (B). In this dataset, selected tracks are also annotated and compared to automated tracking in Table S3 and panel K. C-E, I. Division detections for Oneat (not connected with tracks), TrackMate-Oneat (Oneat divisions connected with TrackMate tracks), TrackMate-Oneat + MARI principle (TrackMate-Oneat with max boundary set for angle between mother cell and daughter cells), and TrackMate “native” track splitting, enabled in TrackMate LAP linking algorithm. F-H, J. Corresponding detection metrics. K. Manually annotated ground truth tracks colorized by the Track ID assigned by automatic tracking used for the experiments.
Oneat model structure. A. Training data annotations in Napari for training a mitosis classifier. B. CNN architecture. Input data is 3 timepoints, 8 x 64 x 64 pixel crop, centered around the annotation ZYX+t centroid. Output is probabilities for classification as mitotic or non-mitotic.