![]()
![]()
This product is a testament to our expertise at KapoorLabs, where we specialize in creating cutting-edge solutions. We offer bespoke pipeline development services, transforming your developmental biology questions into publishable figures with our advanced computer vision and AI tools. Leverage our expertise and resources to achieve end-to-end solutions that make your research stand out.
Note: The tools and pipelines showcased here represent only a fraction of what we can achieve. For tailored and comprehensive solutions beyond what was done in the referenced publication, engage with us directly. Our team is ready to provide the expertise and custom development you need to take your research to the next level. Visit us at KapoorLabs.
BTrack is a TrackMate detector for tracking growing ends of tissue branches using skeletonization implementation of imglib2.
Installation
- Click Help › Update….
- Click the Manage update sites button.
- Select the BTrack update site in the list.
- Click Close and then click Apply changes.
- Restart Fiji.
- The detector would show up in the drop down menu list of TrackMate.
- After the detection, any of the TrackMate tracking algorithms can be used for tracking the skeleton end points.
- Track analysis post tracking can be done using all the functionality available in TrackMate.
Usage
Tissue Detection
BTrack is a tool to analyse the growth of branched tissues. A 2D time-lapse/ 3D image can be analysed with this detector. The file format can be any format readable by Fiji/Bioformats (.tif, .nd2, … ). To run the tracker, select Plugins › Tracking › TrackMate
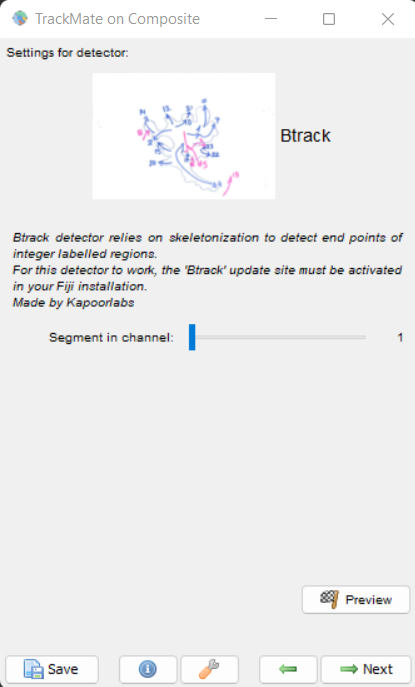
For using this detector select Btrack from the detector menu list of TrackMate. The screen looks as shown here

Detection Panel
Click next and if you are using a hyperstack with Raw and segmentation images in two channels, choose the segmentation image channel for the detector. Click Preview to see the detected end points. The panel for this looks as shown here
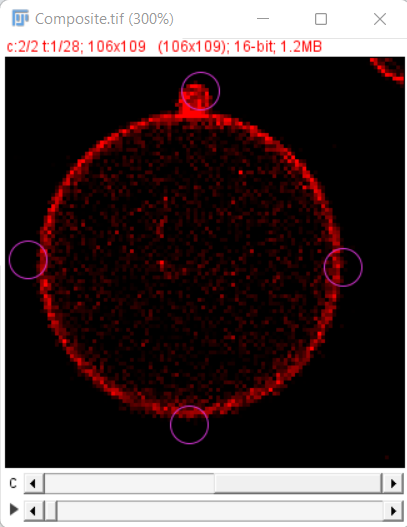
In our example, download from Zenodo we have the Segmentation image in channel 1 and the Raw image in channel 2 and the initial detections look as shown here
Tracking Options
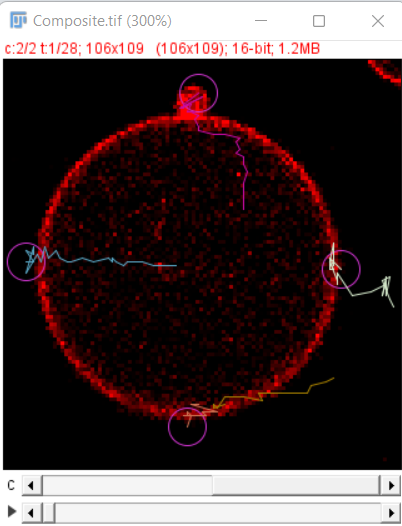
After the initial preview click on the next button to run the detector on all the timepoints, after that you can use the simple LAP tracker of TrackMate 1 to track the growing end points. The final result after tracking looks as shown here
Save Options
All the usual tracking, track editing and saving options that are provided by TrackMate can be used to further analyze and compute the growth rates of individual tracks. For detailed options available for doing so please refer to the manual of TrackMate
Authors
Lead programmer, Mantainer: Varun Kapoor Contributor: Claudia Carabana Garcia
References
-
J. Munkres, “Algorithms for the Assignment and Transportation Problems”, Journal of the Society for Industrial and Applied Mathematics, 5(1):32–38, 1957 March ↩