Scripting with Fiji
Why are scripts useful?
They facilitate reproducible science:
- Document your work
- Automate your analysis
- Share with the world
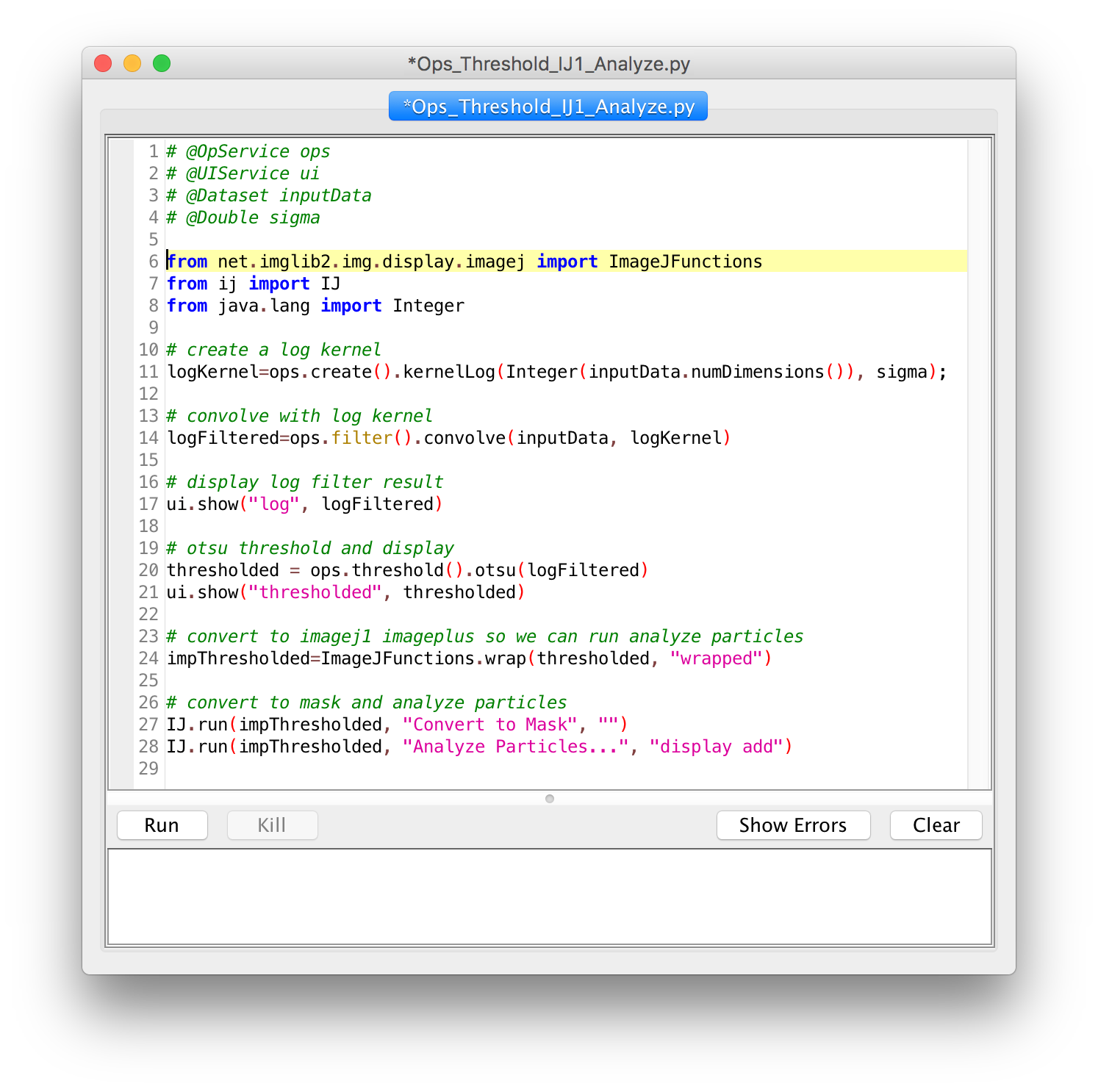
Script Editor
- Comments
- Variables
- Functions
- String manipulation
- Conditionals
- Loops
Macros
Comments
// Comments allow you to put human-readable thoughts
// into your code.
// The goal of this "macro" is simply to teach you about comments!
// Comments help you to remember why you did something:
// Set the value to "2" because my boss said so!
value = 2; // Comments can be added to any line!
// Code can be disabled by commenting it out:
// x = y * 2;
Variables
What is a variable?
- Open "Blobs" image (File > Open Samples > Blobs (25K))
run("Blue");
run("Fire");
Variables
What is a variable?

- Container
- "A piece of code that can vary"
color = "Blue";
run(color);
Variables
// what sorts of values can we assign?
title = "Hello, World!"; // string
intensity = 255; // number
a = exp(x * sin(y)) + atan(x * y – a); // expression
// string constant vs. variable name
text = "title";
text = title;
x = 3;
y = x;
x = 5; // what is the value of y after this?
// the variable is assigned after the expression is evaluated
intensity = intensity * 2;
Strings
name = "copy";
// you can concatenate strings, and strings and numbers
text = "The name is " + name;
// numbers versus strings and more concatenation...
a = 2;
b = 3;
print(a + b);
a = "2";
b = "3";
print(a + b);
// what happens when we run this?
run("Duplicate...", "title=name");
// what's different with this line?
run("Duplicate...", "title=" + name);
Functions

function myFunction(arguments) {
// Your code goes here
}
function setLUT(lutName) {
run(lutName);
}
Functions
print("Hello, world!");
// functions can return values
// Hint: use parameters instead of calling getNumber
number = getNumber("Type in a number!", 5);
// the "run" function is the most important one
// it follows the structure of run("Command...", "Argument(s) String");
run("Duplicate...", "title=New");
// for arguments with spaces, enclose in square brackets
run("Duplicate...", "title=[with spaces]");
Conditionals
#@String instructor
if (getBoolean("Is " + instructor + " going too fast?")) {
hint = "Tell them to to slow down!";
}
else {
hint = "Try to modify the code, play with it...";
}
showMessage("Advice:", hint);
Loops
// For loops:
// They use assignment, test, increment
// Use when you know the # of times the loop will run
for (i = 1; i <= 10; i++) {
print("Counter: " + i);
}
// While loops:
// Use when you do not necessarily know the # of times the loop will run
while (getBoolean("Do you want me to keep going?")) {
print("Ok, I'm still going...");
}
showMessage("Ok, I'm done!");
Tying it all together
Do you notice a bug in this code?
// this example makes a stack of blurred versions of the
// current slice with a range of radii.
radius = getNumber("Maximal radius?", 5);
title = "Blurred stack of " + getTitle();
run("Duplicate...", "title=[" + title + "]");
run("Select All");
run("Copy");
for (i = 1; i <= radius; i++) {
run("Add Slice");
run("Paste");
run("Gaussian Blur...", "radius=" + radius);
}
Macro Recorder
1. Launch the Recorder
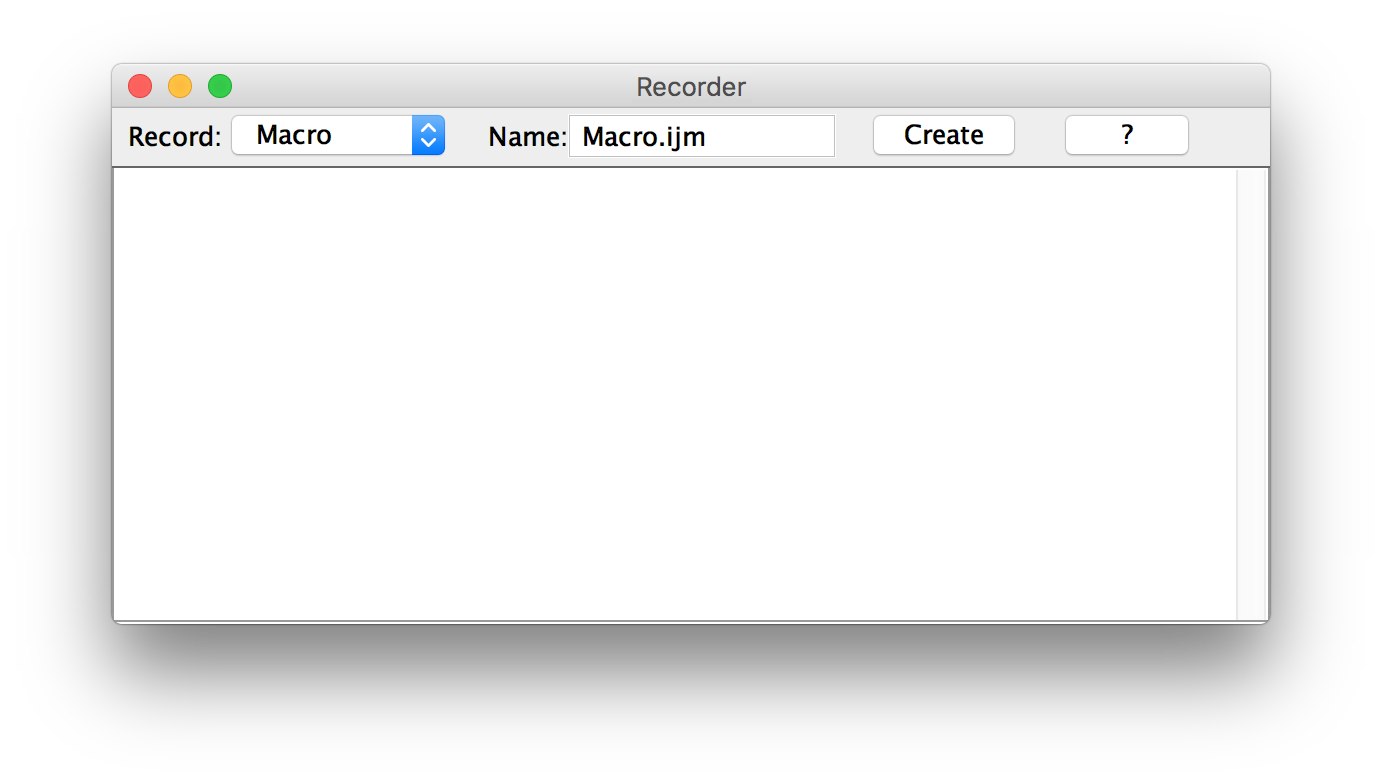
- Use the Search Bar! (Ctrl+L)
- OR: click the Dev icon, then Record...
- OR: Plugins ▶ Macros ▶ Record...
2. Execute operations

Use the Command Finder (Ctrl+L)!
- Open sample images
- Download/unpack "images/C3-jw-30min.zip"
- Plugins ▶ Bio-Formats ▶ Bio-Formats Importer
- Open "C3-jw-30min 5_c5.tif"
- Apply Gaussian blur
- Apply a threshold (Shift+T)
- Create Mask
- Watershed
- Analyze Particles...
- Size: 200-Infinity
- Exclude on edges; Add to Manager
Code Check!
run("Gaussian Blur...", "sigma=2");
setAutoThreshold("Default dark");
//run("Threshold...");
run("Create Mask");
run("Watershed");
run("Analyze Particles...", "size=200-Infinity exclude add");
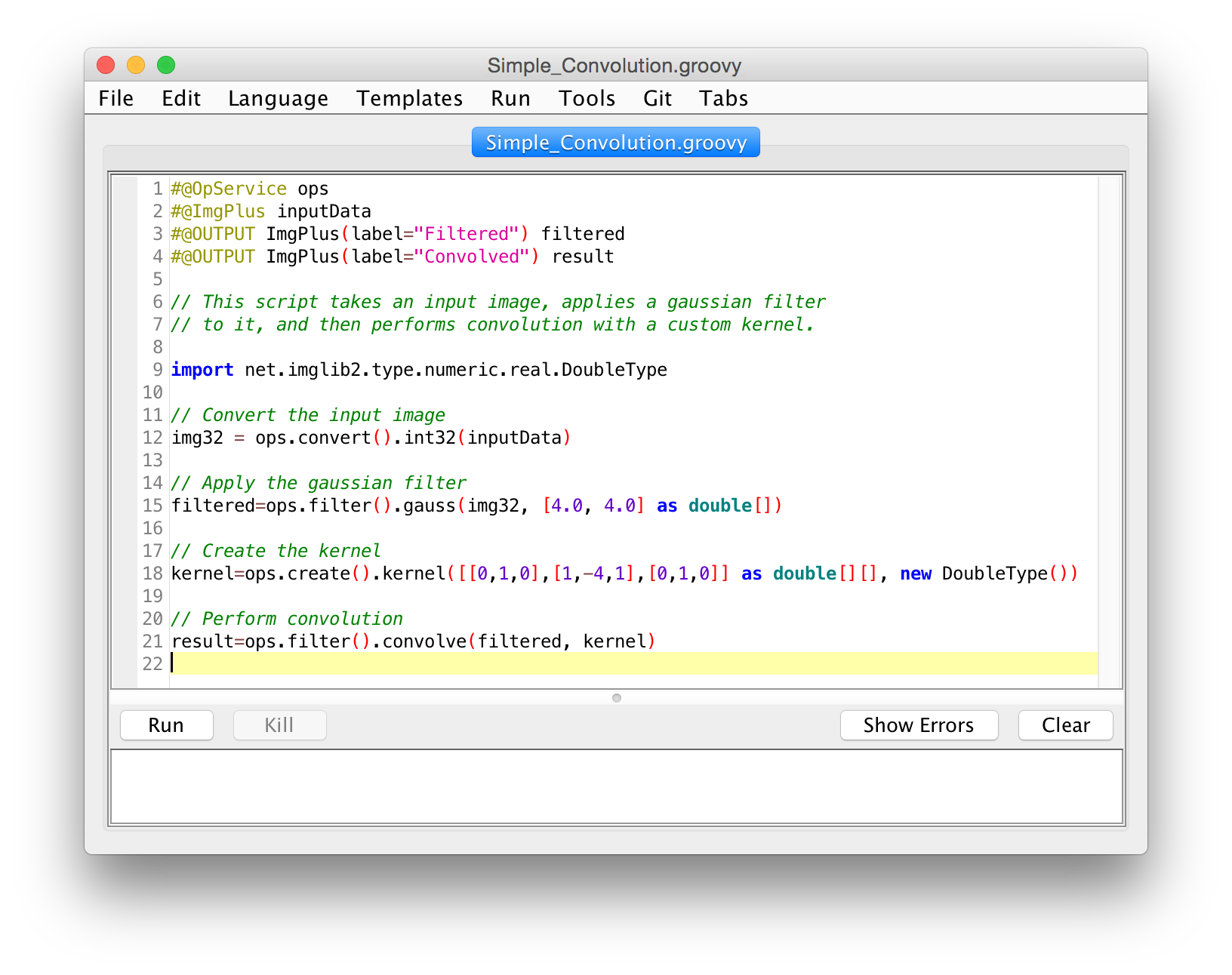
3. Spruce it up
- Click the "Create" button
- Use script parameters
setBatchMode(true)- Use image IDs
- Store in
scripts- In a folder matching the desired menu
- Use underscore for spaces
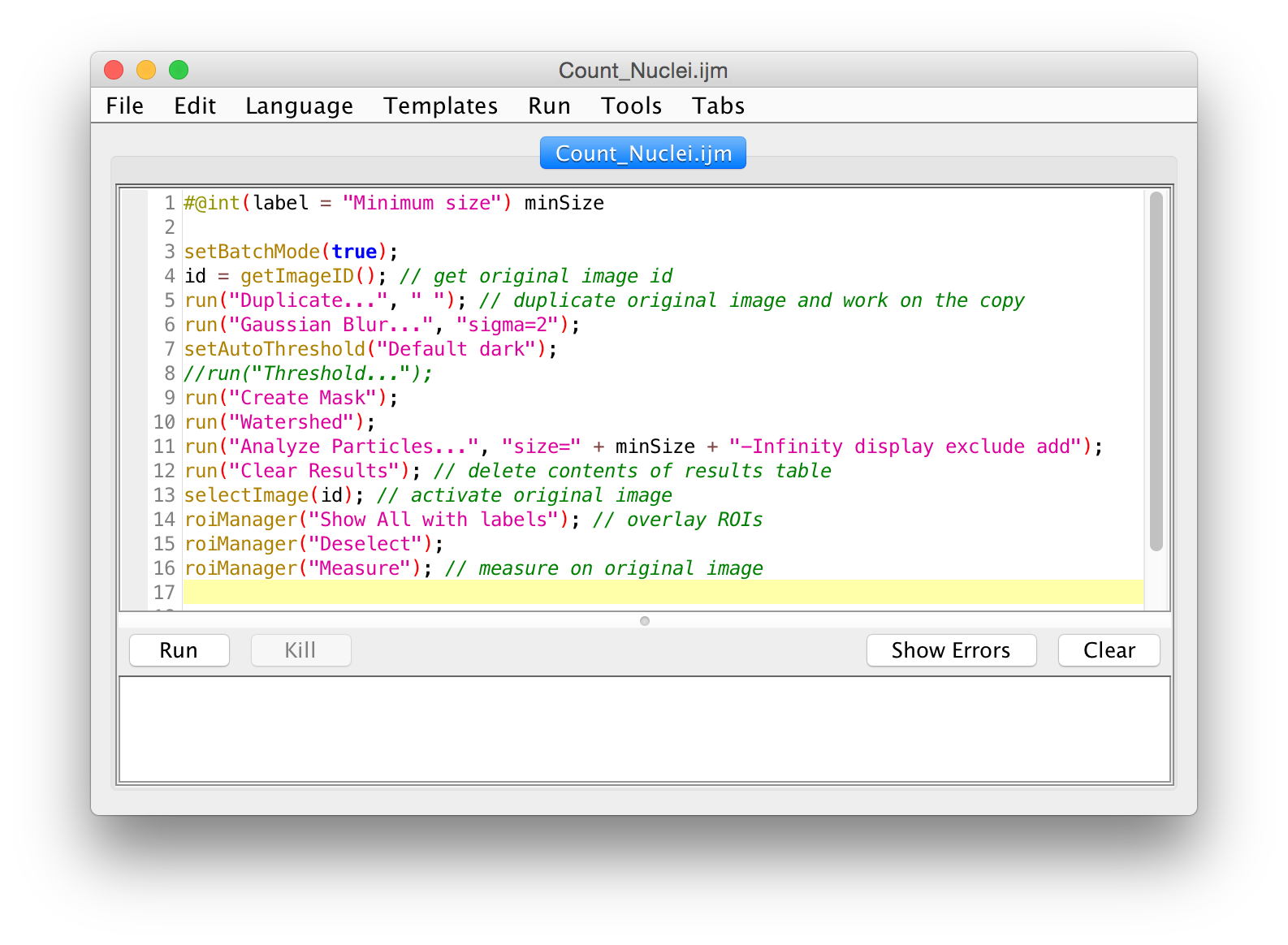
Code Check!
#@int(label = "Minimum size") minSize
setBatchMode(true);
id = getImageID(); // get original image id
run("Duplicate...", " "); // duplicate original image and work on the copy
run("Gaussian Blur...", "sigma=2");
setAutoThreshold("Default dark");
//run("Threshold...");
run("Create Mask");
run("Watershed");
run("Analyze Particles...", "size=" + minSize + "-Infinity display exclude add");
run("Clear Results"); // delete contents of results table
selectImage(id); // activate original image
roiManager("Show All with labels"); // overlay ROIs
roiManager("Deselect");
roiManager("Measure"); // measure on original image
Script parameters

- Templates ▶ Intro ▶ Widgets (BeanShell)
- Run
- Fix the bugs!
- To the
boundedIntegerparameter, add:style="scroll bar" - To the
stringparameter, add:choices={"quick fox", "lazy dog"} - Run again
Batch processing
Process ▶ Batch ▶ Macro...

Templates ▶ ImageJ 1.x ▶ Examples ▶ Process Folder (IJ1 Macro)
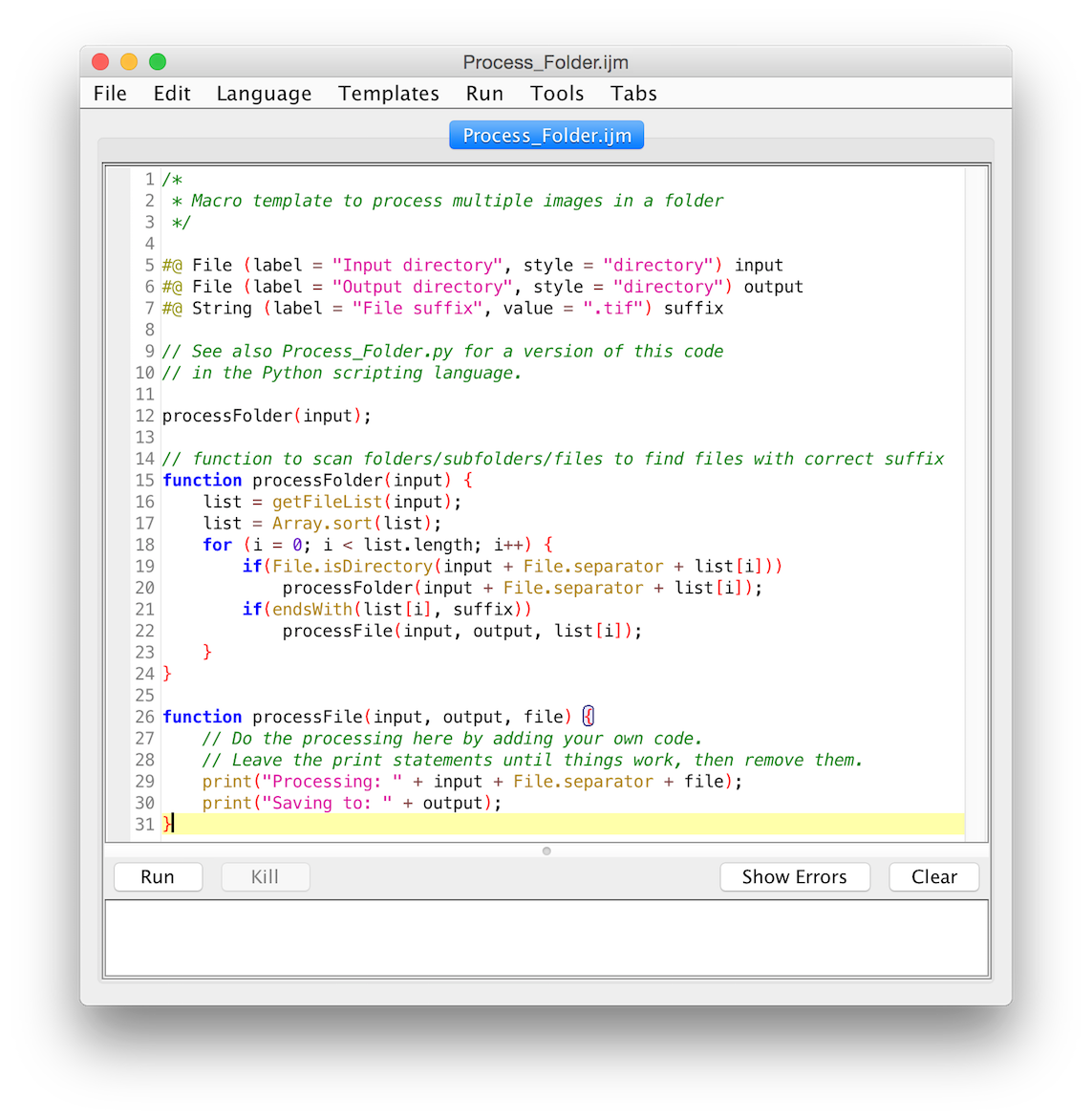
Example script to count nuclei in multiple images in a folder/subfolders:
/*
* Macro to count nuclei in multiple images in a folder/subfolders.
*/
#@File(label = "Input directory", style = "directory") input
#@File(label = "Output directory", style = "directory") output
#@String(label = "File suffix", value = ".tif") suffix
#@int(label = "Minimum size") minSize
processFolder(input);
// function to scan folders/subfolders/files to find files with correct suffix
function processFolder(input) {
list = getFileList(input);
for (i = 0; i < list.length; i++) {
if(File.isDirectory(input + File.separator + list[i]))
processFolder("" + input + File.separator + list[i]);
if(endsWith(list[i], suffix))
processFile(input, output, list[i]);
}
//saves results for all images in a single file
saveAs("Results", output + "/All_Results.csv");
}
function processFile(input, output, file) {
setBatchMode(true); // prevents image windows from opening while the script is running
// open image using Bio-Formats
run("Bio-Formats", "open=[" + input + "/" + file +"] autoscale color_mode=Default rois_import=[ROI manager] view=Hyperstack stack_order=XYCZT");
id = getImageID(); // get original image id
run("Duplicate...", " "); // duplicate original image and work on the copy
// create binary image
run("Gaussian Blur...", "sigma=2");
setAutoThreshold("Default dark");
//run("Threshold...");
run("Create Mask");
run("Watershed");
// save current binary image
save(output + "/Binary_OUTPUT_" + file);
run("Analyze Particles...", "size=" + minSize + "-Infinity exclude add");
selectImage(id); // activate original image
roiManager("Show All with labels"); // overlay ROIs
roiManager("Deselect");
roiManager("Measure"); // measure on original image
// save ROIs for current image
roiManager("Deselect");
roiManager("Save", output+ "/" + file + "_ROI.zip"); // saves Rois zip file
roiManager("Deselect");
roiManager("Delete"); // clear ROI Manager for next image
}
Top tips...
when writing ImageJ Scripts

- Use the Macro Recorder...
- Open the Built-In Macro Functions list
-
print();statements are your friend! - Ask for help on the Forum
Further reading
Help from the community is here:
Scripting guide:
Additional workshops and presentations:

