TreeJ
| TreeJ (ImageJ and Fiji) | |
|---|---|
| Author | Elise Laruelle, Philippe Andrey, Jean-Christophe Palauqui, Alain Trubuil |
| Maintainer | Elise Laruelle |
| File | TreeJ_-1.0.0-SNAPSHOT.jar |
| Source | on GitHub |
| Category | Analysis, Plugins |
This plugin contains an interface to reconstruct interactively and recursively a cell lineage from a static labeled image. It allows to :
- merge two sister cells on the image
- construct a lineage tree displayed on the interface
- annotate the constructed tree
- extract some set of images related to TreeJ functions
- save as a tree file compatible with other tree viewer and/or TreeJ
Usage
Input
TreeJ should be launched on a 2D or 3D labeled image (8 bits, 16 bits or 32 bits). The labeling can be discontinuous and watershed lines, if present, are not a problem. The region labels with zero are considered as the background.
Tree construction
To link two sister cells (in Pair cells panel):
- click on them on the image, it actualizes the corresponding fields (Label1 and Label2)
- or write label numbers in fields Label1 and Label2
Apply to validate the pairing
Reset to put fields in their initial state.
To make a modification (in Unpair or tag panel):
- select the erroneous mother cell with click+Ctrl (or Click+pom on MACOS) and
- or write the label number in the Label field
Validate with the Unpair button.
Tree annotation
- if Tag field is filled during linking action, the created mother cell will have the tag
- select the cell (Ctrl+click| pom+click) or enter its label number in the label field of the unpair and tag panel and enter the tag in the field "Tag"
Apply to validate the tagging.
Image output
- TreeJ doesn't modify the input image. To extract the modified shown image : use Current view
- an image of cells from a Substree
- an image of all cells coming from a mother tagged with the wanted tag
Tree
To saved tree information, two possible formats :
- .treeV home made format, compatible as input of TreeJ. Contains tree and tag information
- .nwk [newick tree], compatible as input of TreeJ, but contains only the tree information
The drawing tree can be exported to a PDF file (named with the input image name) with the Export to PDF button.
Example
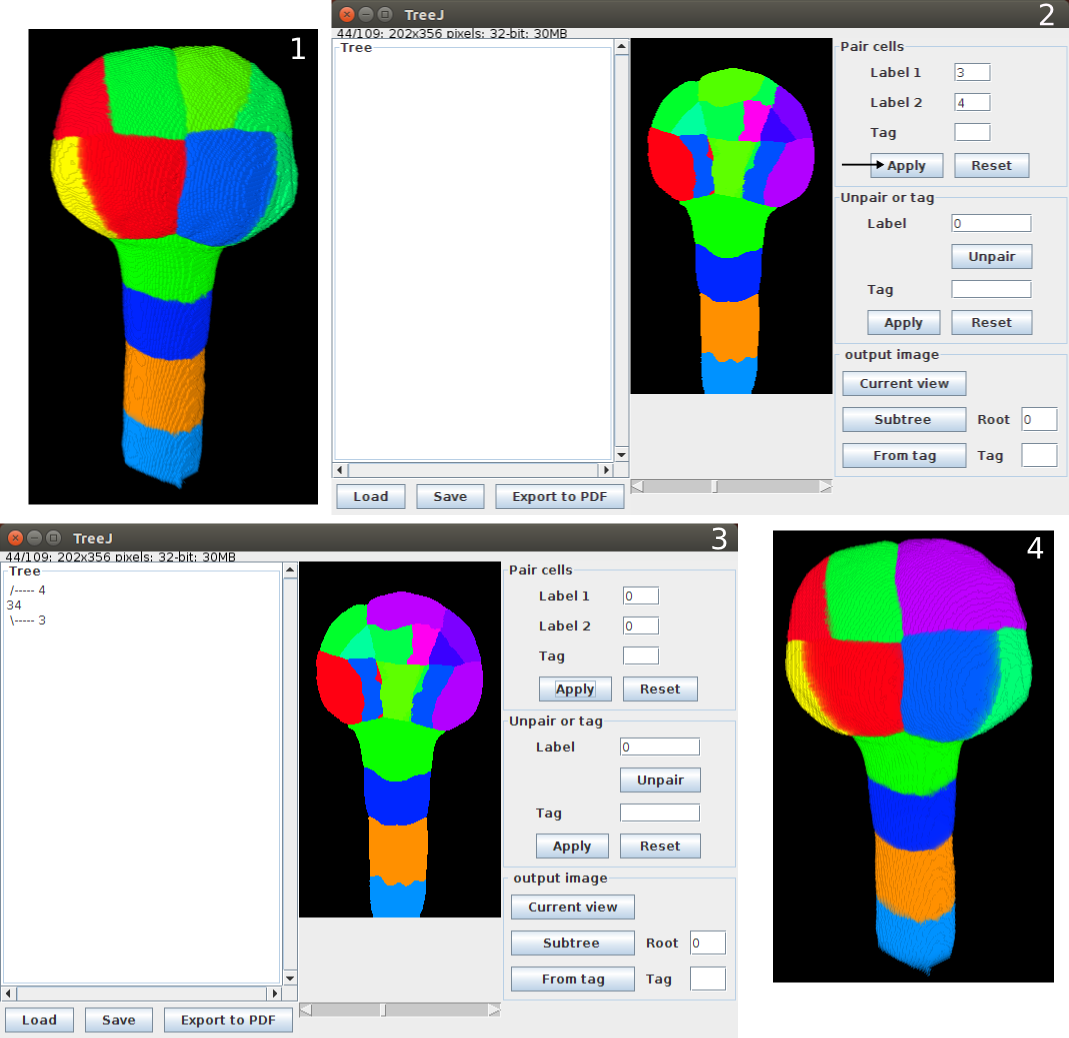
From a segmented image of an Arabidospsi thaliana embryo of 122 cells (FigA), a cell lineage have been backtracked until the 16 cell stage configuration.
- Current view extraction gives the figB, as TreeJ merges sister cells. We see the eight external domains, and there are eight internal domains like shown on the figD (obtain with the external layer and suspensor removing).
- For each domain we can reconstruct the lineage and obtain a tree with TreeJ (see Fig right : a tree of an external apical domain).
- We tagged internal domains of the embryo and extracted the corresponding image them with From Tag (figC).
Installation
Download .jar and put into "plugin" directory of ImageJ or Fiji directory. Reload ImageJ/Fiji and TreeJ will be accessible in the Plugin › TreeJ menu.