Strahler Analysis
| Strahler Analysis (Fiji) | |
|---|---|
| Author | Tiago Ferreira |
| Maintainer | Tiago Ferreira |
| File | hIPNAT_.jar (Neuroanatomy update site) |
| Source | on GitHub |
| Initial release | April 2016 |
| Category | Plugins, Neuroanatomy, Analysis, Skeleton |
A plugin from the Neuroanatomy update site that performs Strahler analysis on topographic skeletons (2D/3D). Strahler numbering is a numerical procedure that summarizes the branching complexity of mathematical trees.
Contents
Description
The analysis occurs through progressive pruning of terminal branches, iterative tree simplification, a method that requires detecting all terminal branches (i.e., branches that contain an end-point) and all the degree-one paths leading to them.
Strahler Analysis takes a binary or 8-bit grayscale image (2D or 3D) containing a single arbor, and calls AnalyzeSkeleton iteratively to retrieve Horton-Strahler numbers from the skeletonized centerlines of the input image. Each iteration includes three operations: 1) a (re)-skeletonization step to ensure that arbor remains represented by its centerlines, 2) an elimination step in which terminal-branches are pruned from the image and 3) an analysis step in which pruned branches are counted and measured. The iteration ceases as soon as all branches have been eliminated or a unresolved closed loop has been detected in the pruned arbor.
Parameters
- Tree Classification
-
- Infer root end-points from rectangular ROI
- This option is only available when a rectangular ROI is present. It is described in Non-radial arbors.
- Ignore single-point arbors (Isolated pixels)
- Elimination of end-point branches may give rise to single point arbors. Such 'debris' have 1 end-point but no slab branches or junctions. When this option is selected, single-point arbors will be discarded on each iteration. If deselected, the total number of end-points may be overestimated.
- Elimination of Skeleton Loops
-
- Method
- Strahler Analysis cannot process skeletons containing closed loops and will output a warning message when such structures have been detected. The available methods in this drop-down menu define how closed loops should be resolved by and are described in the AnalyzeSkeleton documentation page.
- Unsegmented image
- The initial non-thinned image to be used by AnalyzeSkeleton for intensity-based elimination of closed loops. This option is only used if either Lowest intensity voxel or Lowest intensity branch is chosen as Method. Note that if an intensity-based method is selected but the chosen image is a binary one, closed loops will not be resolved.
- Output Options
-
- Display Iteration stack
- If checked, an image stack that documents individual pruning cycles will be displayed. End-points and Junction-points positions are appended to the stack.
- Show detailed information
- If checked, analysis will run in verbose mode by outputting detailed measurements and by logging debug messages.
Root Detection
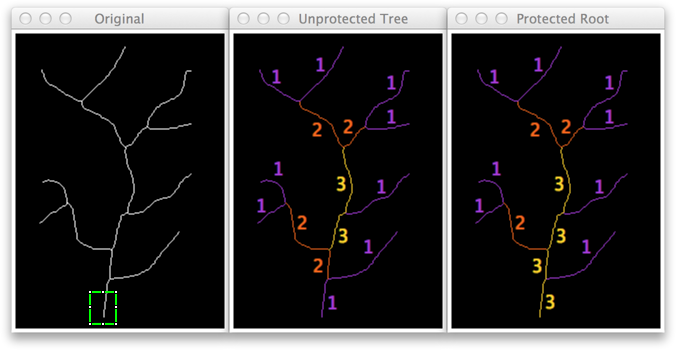
The problem with undiscriminated elimination of terminal branches is that a root-branch containing an end-point is always eliminated on the first iteration step. In order to protect root branches from elimination, Strahler Analysis needs to know where root branches are located. As of version 1.4.0, root-detection is implemented by means of a rectangular ROI containing the root branch and by activating the Infer root end-points from rectangular ROI option. Here is an example:
Notes on Root Detection
- Only a rectangular ROI can be used to mark the root branch. This is intentional: The way the root-detection algorithm works is by protecting all end-points that are contained by the ROI from end-point elimination. Using complex ROIs (e.g., discontinuous or containing internal holes) would make this task much more cumbersome.
- The ROI only needs to contain root end-point(s) and it should not matter if its boundaries intercept other branches. However, measurements on root-branches may be inaccurate if the ROI contains junction(s) points. The best way to ensure the algorithm ran as expected is to visually inspect all the slices in the Iteration stack.
- Root detection may not be required in the case of radial arbors (i.e., tree-like structures that branch out evenly in multiple directions), if root(s) remain connected in the center of the arbor (as in animation above). In neurobiology, radial-ramification is an anatomical hallmark of certain cell types such as Retinal Ganglion Cells, Chandelier neurons or Drosophila Class IV sensory neurons.
- If you are batch processing multiple images you should work with .tif files: When saving as TIFF, ImageJ will store the active ROI in the image header, making it immediately available when the image is open.
Results
The plugin produces three types of outputs:
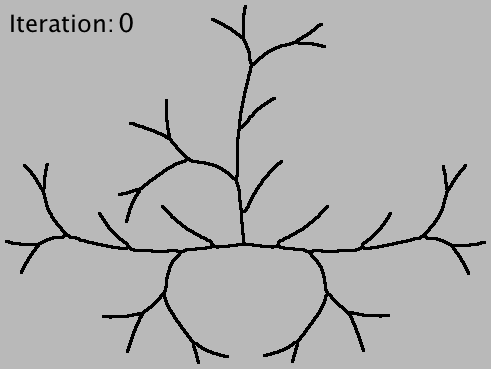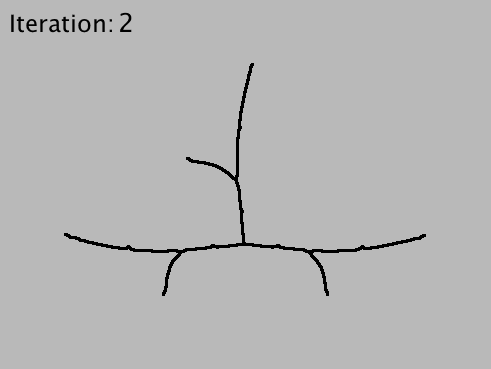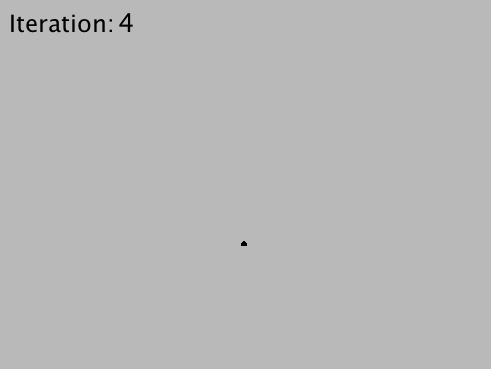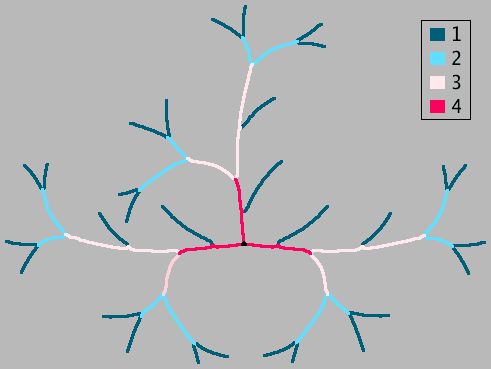
- Strahler Image
- A heat-map image, in which branches are color-coded by their Horton-Strahler numbers. By default, a calibration ramp (Analyze › Tools › Calibration Bar...) is added as an overlay. WYSIWYG versions (RGB images) of this Strahler Color Map image can be obtained by pressing ⇧ Shift+F (shortcut for Image › Overlay › Flatten).
- Strahler table
- Table listing Horton-Strahler counts. The extension and format of this Strahler Table can be specified in Edit › Options › Input/Output.... It contains the following data:
- # End-point Branches
- The number of branches for each Horton-Strahler order.
- Ramification ratios
- Ramification or bifurcation ratios are the quotients between branches of consecutive orders. An overall ratio may be obtained by averaging ratios across orders.
- Iteration log
- If Show detailed information is checked, Average branch length, N. of trees, N. of branches, N. of junctions, N. of triple points, N. of quadruple points are also retrieved for each iteration. These are described in the AnalyzeSkeleton's documentation page.
Installation
To install Strahler Analysis you must use Java 8 and subscribe to the Neuroanatomy update site.
Related Links
- AnalyzeSkeleton and Skeletonize3D, analysis of topographic skeletons
- Sholl Analysis, bitmap morphometry based on the Sholl technique
References
Original publications by Robert E. Horton and Arthur N. Strahler:
- Horton, R. E. (1945), "Erosional development of streams and their drainage basins: hydro-physical approach to quantitative morphology", Geological Society of America Bulletin 56 (3): 275–370, DOI 10.1130/0016-7606(1945)56[275:EDOSAT]2.0.CO;2.
- Strahler, A. N. (1952), "Hypsometric (area-altitude) analysis of erosional topology", Geological Society of America Bulletin 63 (11): 1117–1142, DOI 10.1130/0016-7606(1952)63[1117:HAAOET]2.0.CO;2.
- Strahler, A. N. (1957), "Quantitative analysis of watershed geomorphology", Transactions of the American Geophysical Union 38 (6): 913–920 (PDF).
Citing
Plugins from the Neuroanatomy update site:
Skeletonization and Skeleton Analysis:
- Ignacio Arganda-Carreras, Rodrigo Fernandez-Gonzalez, Arrate Munoz-Barrutia, Carlos Ortiz-De-Solorzano, "3D reconstruction of histological sections: Application to mammary gland tissue", Microscopy Research and Technique, Volume 73, Issue 11, pages 1019–1029, October 2010.
- Michael Doube, Michal M. Klosowski, Ignacio Arganda-Carreras, Fabrice P. Cordelieres, Robert P. Dougherty, Jonathan S. Jackson, Benjamin Schmid, John R. Hutchinson, Sandra J. Shefelbine, BoneJ: Free and extensible bone image analysis in ImageJ, Bone, Volume 47, Issue 6, December 2010, Pages 1076-1079.
Acknowledgments
To all the developers of AnalyzeSkeleton.
License
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.